Corrigendum: Improving anti-tumor efficacy of low-dose Vincristine in rhabdomyosarcoma via the combination therapy with FOXM1 inhibitor RCM1
- 1Division of Pulmonary Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 2The Materials Science and Engineering Program, College of Engineering and Applied Science, University of Cincinnati, Cincinnati, OH, United States
- 3Center for Lung Regenerative Medicine, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 4Division of Pulmonary and Critical Care and Sleep Medicine, Department of Internal Medicine, University of Cincinnati, Cincinnati, OH, United States
Rhabdomyosarcoma (RMS) is a highly metastatic soft-tissue sarcoma that often develops resistance to current therapies, including vincristine. Since the existing treatments have not significantly improved survival, there is a critical need for new therapeutic approaches for RMS patients. FOXM1, a known oncogene, is highly expressed in RMS, and is associated with the worst prognosis in RMS patients. In the present study, we found that the combination treatment with specific FOXM1 inhibitor RCM1 and low doses of vincristine is more effective in increasing apoptosis and decreasing RMS cell proliferation in vitro compared to single drugs alone. Since RCM1 is highly hydrophobic, we developed innovative nanoparticle delivery system containing poly-beta-amino-esters and folic acid (NPFA), which efficiently delivers RCM1 to mouse RMS tumors in vivo. The combination of low doses of vincristine together with intravenous administration of NPFA nanoparticles containing RCM1 effectively reduced RMS tumor volumes, increased tumor cell death and decreased tumor cell proliferation in RMS tumors compared to RCM1 or vincristine alone. The combination therapy was non-toxic as demonstrated by liver metabolic panels using peripheral blood serum. Using RNA-seq of dissected RMS tumors, we identified Chac1 as a uniquely downregulated gene after the combination treatment. Knockdown of Chac1 in RMS cells in vitro recapitulated the effects of the combination therapy. Altogether, combination treatment with low doses of vincristine and nanoparticle delivery of FOXM1 inhibitor RCM1 in a pre-clinical model of RMS has superior anti-tumor effects and decreases CHAC1 while reducing vincristine toxicity.
Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children, accounting for about 350 cases in the United States each year (1–5). RMS is highly metastatic, and these patients have a poor prognosis, with the 5-year survival rate being around 20% (2–4). Chemotherapy is often used to treat these patients, such as anthracycline-based agents like doxorubicin or epirubicin, alkylating agents like ifosfamide and cyclophosphamide, cytotoxic therapeutics like dactinomycin, and vincristine (VCR), a vinca alkaloid agent (6). VCR is a cell-cycle specific therapeutic that has been widely accepted as a preferred initial, recurrent, and metastatic treatment for RMS patients (2, 7, 8). Unfortunately, many chemotherapies including VCR are toxic, leading to weight loss, decreased blood counts, and neuropathy (2, 5, 8, 9). VCR is used in combination with other therapeutics to combat toxicity, while also increasing efficacy (5, 7). Despite this, RMS is highly recurrent and will often develop therapeutic resistance to these drugs (2, 3), which indicates an urgent need for new therapies.
FOXM1, a well-known oncogene, is an important transcription factor that is crucial to many pathways and processes, most notably proliferation, mitosis, and cell survival (10–14). FOXM1 is overexpressed in many cancers, including RMS, and the high-levels of FOXM1 are clinically associated with a worse prognosis in RMS patients (15, 16). FOXM1 is primarily expressed during embryogenesis in the developing tissue, with minimal-to-no expression in normal healthy tissue, making it a very attractive therapeutic target during carcinogenesis (17–20). Previous studies have shown that the loss of FOXM1 in tumor cells results in decreased tumor cell proliferation, metastasis, angiogenesis, and increased tumor cell death (21–24). However, many of the targeted approaches to inhibit FOXM1 have largely been unsuccessful due to their lack of specificity and off-target effects, including toxicity (17, 23).
Recently, we have identified a small molecule inhibitor of FOXM1 (Robert Costa Memorial Drug-1, RCM1) that has shown to be an effective anti-tumor agent and is also non-toxic in pre-clinical mouse models (19, 25). RCM1 removes FOXM1 from the nucleus into the cytoplasm, leading to FOXM1 ubiquitination and proteasomal degradation (25). Treatment with RCM1 reduces tumor cell proliferation, migration and colony formation while also inducing apoptosis across several cancer types, including RMS (19). RCM1 treatment increases duration of mitosis in tumor cells (19), suggesting that the use of RCM1 can synergize with anti-mitotic chemotherapeutic agents to increase the efficacy of anti-cancer treatment. Furthermore, dual-treatment with RCM1 and anti-mitotic drugs provides an opportunity to decrease the drug concentrations and decrease side effects of anti-cancer therapy as a result. In the present studies, we investigated the effect of RCM1 in combination with low dose of VCR in RMS mouse models. Our results demonstrate a combinatorial effect between RCM1 and VCR to increase anti-tumor efficacy on RMS tumors, potentially suggesting a novel therapeutic option for RMS patients.
Materials and methods
Cell lines and reagents
Human RD (ATCC) is an embryonal RMS cell line derived from a 7-year-old Caucasian female previously treated with cyclophosphamide and radiation. RD is treatment refractory (26), has MYC amplification (27), mutations in NRAS and TP53 (28), and is sensitive to VCR (29). Rd76-9 is a murine derived embryonal RMS cell line, isolated from a methylcholanthrene-induced mouse RMS tumor in a female C57BL/6 mouse (30) and was provided by Dr. Tim Cripe (Nationwide Children’s Hospital, Columbus, OH). The small molecule compound RCM1 (2-[2-oxo-2-(thiophen-2-yl) ethyl]sulfanyl -4,6-di(thiophen-2-yl)pyridine-3-carbonitrile) was synthesized by Vitas-M Laboratory (95% purity). Vincristine Sulfate (VCR, NDC 61703-309-16) was purchased through in-house pharmacy at Cincinnati Children’s Hospital.
Nanoparticle synthesis
Bisphenol A glycerolate diacrylate, 4,4’-Trimethylenedipiperidine, 6-amino-1-hexanol, Oleic acid, polyethylenimine (PEI, Mn ~600) poly(ethylene glycol) (PEG, Mn ~2000), and Folic acid (FA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Lecithin (from Soybean) was purchased from Tokyo Chemical Industry Co., Ltd. (TCI). DyLight 650 or 800 NHS ester, carbodiimide hydrochloride (EDC), N- Hydroxysuccinimide (NHS), SnakeSkin™ Dialysis Tubing, and 10K MWCO were purchased from ThermoFisher Scientific (Waltham, MA, USA).
The amphiphilic poly-beta amino ester (aPBAE) nanoparticle backbone was synthesized via a Michael Addition. Briefly, the Bisphenol A glycerolate diacrylate was first mixed with 4,4’-Trimethylenedipiperidine in DMSO at 50°C for 24 hours. The mixture was then added to 6-amino-1-hexanol with the temperature increased to 90°C and held for another 24 hours. The polymer backbone was capped by a FA modified PEI. The FA and PEG modifications were processed via EDC/NHS coupling, as described previously (31, 32). To encapsulate RCM1, nanoparticle components were mixed with RCM1 in DMSO, then moved to an aqueous condition to allow DMSO to diffuse and the nanoparticles to assemble for 4 hours, followed by dialysis for 48 hours to remove DMSO and impurities. UV/Vis spectroscopy has been widely used to determine drug loading capacity using various solvents, including DMSO (33–35), so RCM1-encapsulated nanoparticles were characterized by using UV/Vis spectroscopy to determine the amounts of RCM1 according to the standard curve for estimation of encapsulation concentration.
Mouse models
C56Bl/6J mice were purchased from the Jackson laboratory. To generate the subcutaneous syngeneic murine model, 1x106 Rd76-9 rhabdomyosarcoma cells were re-suspended in equal volumes of PBS : Matrigel (Corning) and were injected into the flanks of 8-12 weeks old C57Bl/6J mice. Animals were treated with VCR or saline (control) every 7 days via intraperitoneal injections. Animals were treated with RCM1 encapsulated nanoparticles or empty nanoparticles (control) every 48 hours intravenously via the eye vein. Tumors were measured using calipers, and volumes were calculated in cubic millimeters using where L is the largest diameter and W is the diameter perpendicular to L. Serum was collected from animals treated with empty nanoparticles, treated with 0.25mg/kg VCR alone, 8µg RCM1 nanoparticles alone or treated with combination of 0.25mg/kg VCR and 8µg RCM1 nanoparticles. Each animal study had 3-7 mice per group.
In vitro growth curve analysis
3x105 Tumor cells per well were seeded in 6-well plates and allowed to grow for 24 hours or 48 hours. After 24 hours, tumor cells were then treated with RCM1, VCR or RCM1 + VCR for 24 hours. Trypan blue was used to exclude dead cells and viable cells were counted using a Hemocytometer. Experiments were performed in triplicate.
Immunostaining
3x105 Tumor cells per well were seeded on 24mm square cover slips in 6-well plates and allowed to grow for 24 hours. Tumor cells were then treated with RCM1, VCR or RCM1 + VCR for 24 hours. Tumor cells were then fixed and stained as previously described (36). To visualize the nucleus, Hoechst 33342 (ThermoFisher Scientific) was used as a counter stain. For quantification, 5 random fields were acquired per sample and quantified using ImageJ. To perform immunostaining of tumor tissue, paraffin embedded Rd76-9 subcutaneous tumor sections were stained as described previously (37). 5 Random fields per sample were acquired and quantified using ImageJ. Antibodies used for immunostaining were anti-Ki-67 (Invitrogen, MA5-14520), anti-phospho-histone H3 (Santa Cruz, sc-374669 (C-2)), anti-CD31 (R&D, AF3628), anti-Cleaved-Caspase 3 (R&D, MAB835), anti-CHAC1 (Novus Biologicals, OTI1E2).
qRT-PCR and western blot
For stable knockdown of CHAC1, Rd76-9 cells were transduced with GIPZ Chac1 shRNA (V3LMM_499697, Horizon). GIPZ non-silencing lentiviral shRNA (RHS4346, Horizon) was used as control. Lysis Buffer (Qiagen) and β-Mercaptoethanol were used to lyse cells in vitro. Taqman gene expression assays for Chac1 and Beta-Actin were purchased from ThermoFisher. qRT-PCR was performed as previously described (38, 39). Protein extracts were prepared as described (40, 41). Antibodies used for western blots were anti-CHAC1 (Sigma, AV42623), and anti-Vinculin (Cell Signaling, E1E9V).
RNA-seq and data processing
Dissected Rd76-9 tumors from control (treated with empty nanoparticle and saline), 0.25mg/kg VCR, 8µg RCM1 and 0.25mg/kg VCR + 8µg RCM1 groups were snap frozen in liquid nitrogen 16 days post tumor inoculation. RNA was extracted from bulk tumor samples and were sent for sequencing. Quality of RNA was determined using Fragment Analyzer with an average RQN for all samples of 9.7. RNA libraries were prepared for all samples using Illumina TruSeq Stranded mRNA Prep to generate poly-A enriched, non-stranded RNA libraries. Sequencing was performed using NoveSeq6000 with an estimated 30 million read per sample. Reads were aligned to the GRCm38 mouse genome and quantified using an index transcriptome version of GRCm38 using Kallisto using standard settings (42). Raw counts were normalized using DESeq2 (43). Differential gene expression between conditions was performed using DEseq2 which uses a negative binomial model for each gene. The Wald test was used for hypothesis testing when comparing the two groups. All p-values attained were corrected for multiple testing using the Benjamini and Hochberg method which is the default method in DEseq2. In the standard DESeq2 algorithm, alpha for false-discovery rate is set to 0.1 by default. Heatmap was generated using the pheatmap R package and the volcano plot was generated using the EnhancedVolcano R package (https://github.com/kevinblighe/EnhancedVolcano). Venn diagrams for differentially expressed genes were created using AltAnalyze (44).
Statistical analysis
The Student t test, one-way ANOVA, one-way ANOVA RM and two-way ANOVA RM were used to determine statistical significance. P-values <0.05 were considered significant. Values were shown as mean SD. All statistical analyses were obtained using GraphPad Prism.
Study approval
All animal studies were approved by Cincinnati Children’s Research Foundation Institutional Animal Care and Use Committee and covered under our animal protocol (IACUC2022-0041). The Cincinnati Children’s Research Foundation Institutional Animal Care and Use Committee is an AAALAC and NIH accredited institution (NIH Insurance #8310801).
Results
VCR and RCM1 synergize in murine RMS cells in vitro
To determine the half maximal inhibitory concentrations (IC50) of VCR and RCM1, dose response curves were generated for each single agent. After 24 hours, both VCR and RCM1 reduced the number of Rd76-9 murine RMS cells in a dose-dependent manner (Figure 1A). Using the dose response curves for VCR and RCM1, we selected IC50 concentrations of 2.1nM for VCR and 8.7µM for RCM1 to use in combination treatment. Using these concentrations, the dual treatment with low doses of VCR and RCM1 synergized to reduce the number of Rd76-9 tumor cells after 24 hours compared to the single agents and control (Figure 1B, S1A). To further characterize this novel dual-drug therapy, we performed immunostaining of tumor cells using various markers. Since both RCM1 and VCR are cell-cycle specific anti-tumor agents, we first explored the effects of the combination therapy on tumor cell proliferation. Cell proliferation was assessed using immunostaining for Ki-67, a general proliferation marker, and phospho-histone H3 (PH3), a mitosis specific marker. After 24 hours, the combination therapy synergized to reduce the number of Ki-67-positive cells compared to each single agent (Figure 1C). Also, the two agents synergized to reduce the number of mitotic cells after 24 hours (Figure 1D). To characterize the effects of this combination therapy on cell death, we performed immunostaining for Cleaved-Caspase 3, a well-known marker for apoptosis. After 24 hours, this combination therapy synergized to increase the number of apoptotic RMS cells, suggesting that the dual-drug therapy with low doses of RCM1 and VCR induces tumor cell death with higher efficiency than single agents alone (Figure 1E). These results demonstrate that RCM1 in combination with VCR synergize to inhibit proliferation and increase apoptosis of murine RMS tumor cells in vitro.
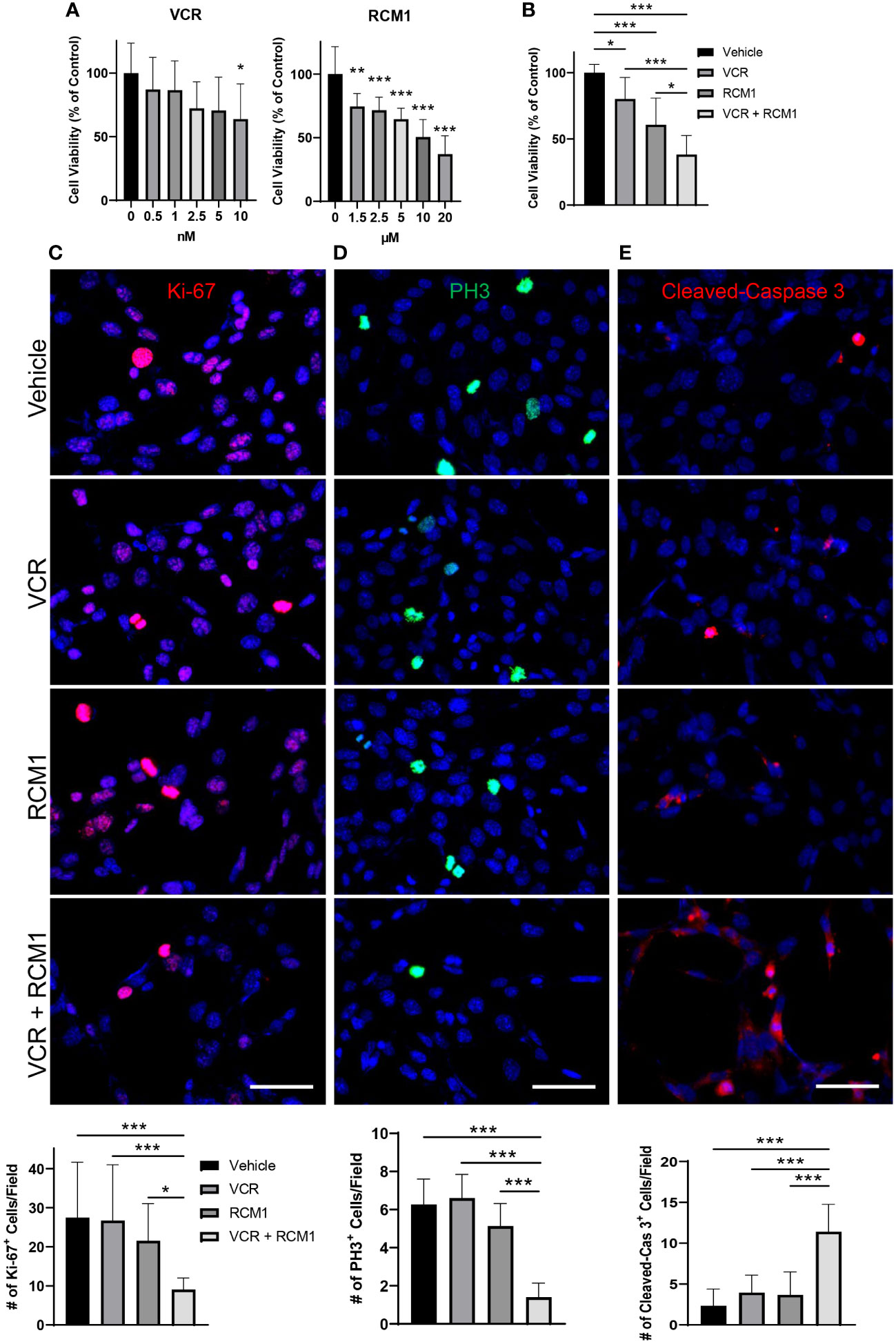
Figure 1 Combination of VCR and RCM1 is more efficient than single agents in reducing growth of murine Rd76-9 rhabdomyosarcoma cells in vitro. (A), Left: Vincristine (VCR) reduced Rd76-9 tumor cell viability in a dose-dependent manner after 24 hours of treatment compared to vehicle (saline) control. Right: RCM1 reduced Rd76-9 tumor cell viability in a dose-dependent manner after 24 hours of treatment compared to vehicle (DMSO) control. Graphs show the percentage of live tumor cells per group compared to vehicle control. (B), Combination therapy using IC50 concentrations of VCR (2.1nM) and RCM1 (8.7µM) reduced Rd76-9 tumor cell viability compared to single agents at IC50 concentrations and to vehicle (saline + DMSO). The graph shows the percentage of live tumor cells per group compared to vehicle control after 24 hours of treatment. (C), Combination therapy decreased Rd76-9 tumor cell proliferation (Ki-67, red) and mitosis (D, PH3, green), and increased apoptosis (E, Cleaved-Caspase 3, red) compared to single agents or vehicle control after 24 hours of treatment. 5 random fields per sample were used to quantify the number of Ki-67+, PH3+ and Cleaved-Caspase 3+ cells per group. Values are shown as mean ± SD. *P<0.05; **P<0.01; ***P<0.001. Scale bar=50µm.
VCR and RCM1 synergize in human RMS cells in vitro
Since the dual treatment with low doses of RCM1 and VCR was effective in murine RMS cells, we investigated the effects of this combination therapy using a human derived RMS cell line, RD. To determine an optimal treatment dose, we generated the dose response curves for each drug and selected 3.5nM for VCR and 3.0µM for RCM1 to use in combination therapy (Figure 2A). Consistent with the mouse data, this dual therapy with low doses of VCR and RCM1 synergized to reduce the number of human RD tumor cells more efficiently compared to single agents (Figure 2B, S1B). Next, we assessed tumor cell proliferation by performing immunostaining for Ki-67 and PH3. Similar to the murine RMS cells, the combination therapy synergized to reduce the number of Ki-67-positive proliferating tumor cells (Figure 2C) and PH3-positive mitotic tumor cells compared to single agents (Figure 2D). Importantly, combination of low doses of VCR and RCM1 synergized to increase the number of apoptotic tumor cells as shown by immunostaining for Cleaved-Caspase 3 (Figure 2E). Taken together, these data suggest that the RCM1 and VCR combination therapy can be a promising effective therapy for RMS in vivo.
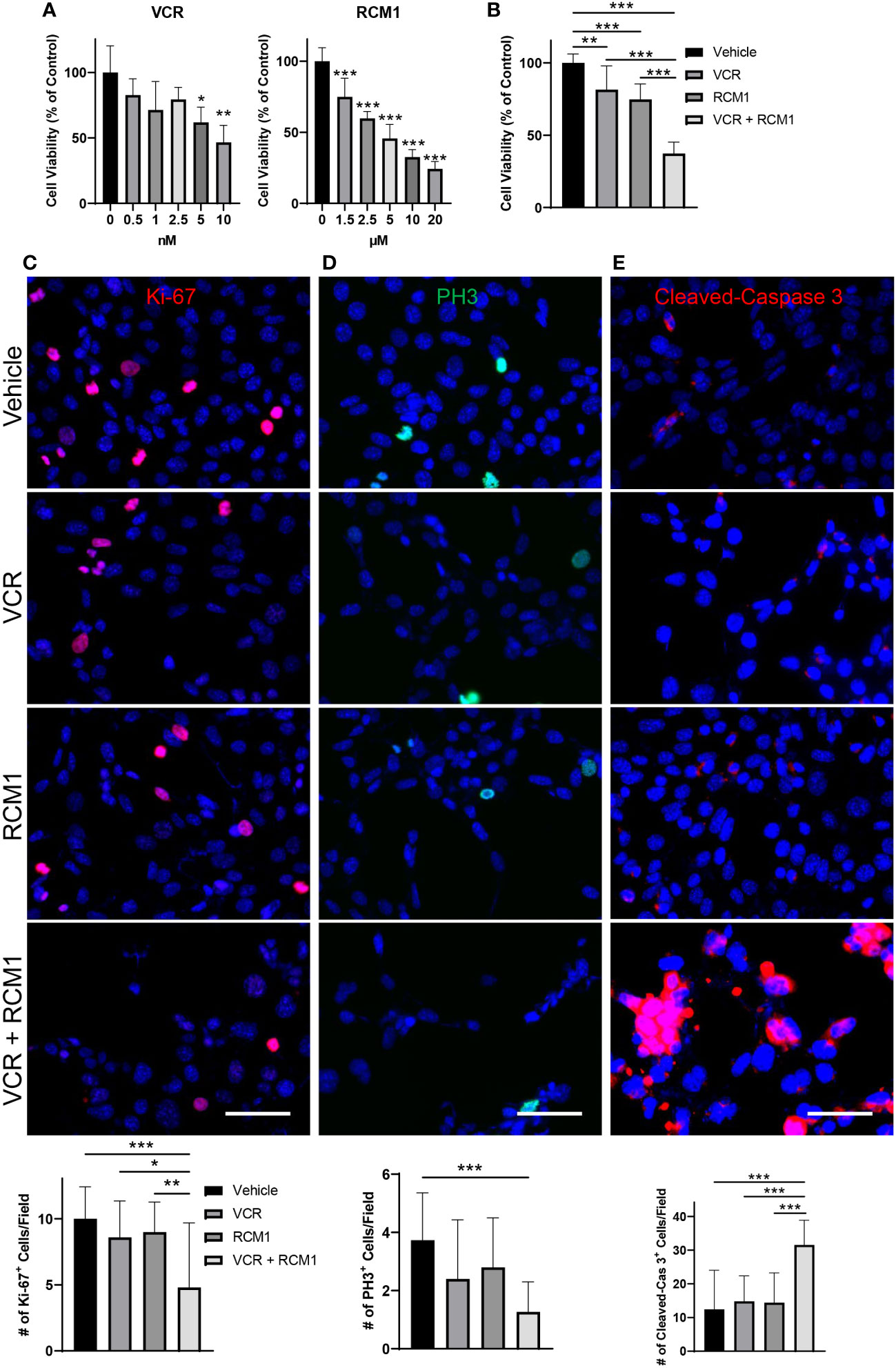
Figure 2 The combination of VCR and RCM1 is more efficient than single agents in reducing growth of human RD rhabdomyosarcoma cells in vitro. (A), Left: VCR reduces RD tumor cell viability in a dose-dependent manner after 24 hours of treatment compared to vehicle (saline) control. Right: RCM1 reduces RD tumor cell viability in a dose-dependent manner after 24 hours of treatment compared to vehicle (DMSO) control. Graphs show the percentage of live cells per group compared to vehicle control. (B), Combination therapy using IC50 concentrations of VCR (3.5nM) and RCM1 (3.0µM) reduced tumor cell viability compared to single drugs or vehicle (saline + DMSO) control after 24 hours of treatment. The graph shows the percentage of live cells per group compared to vehicle control. (C), Combination therapy reduces proliferation (Ki-67, red) of RD tumor cells compared to single drugs or vehicle control after 24 hours of treatment. (D), Combination therapy reduces mitosis (PH3, green) of RD tumor cells compared to vehicle control after 24 hours of treatment. (E), Combination therapy induces apoptosis (Cleaved-Caspase 3, red) of RD tumor cells compared to single drugs alone or vehicle control after 24 hours of treatment. 5 random fields per sample were used to quantify the number of Ki-67+, PH3+ and Cleaved-Caspase 3+ cells per group. Values are shown as mean ± SD. *P<0.05; **P<0.01; ***P<0.001. Scale bar=50µm.
Treatment with either VCR or RCM1 alone reduces tumor burden in a mouse model of RMS
To determine the half maximal inhibitory concentrations (IC50) of VCR and RCM1 as single agents in vivo, Rd76-9 murine RMS cells were injected subcutaneously into the flanks of C57Bl/6J mice to create a syngeneic RMS model. Mice were treated with VCR at days 7 and 14 after tumor cell inoculation (Supplementary Figure S2A). Treatment with VCR reduced tumor volume in a dose-dependent manner (Supplementary Figure S2B). Using tumor volumes measured at day 16, we generated a dose response curve to determine the IC50 concentration for VCR (0.25mg/kg) that later would be used for the combination therapy with RCM1 (Supplementary Figure S2C). Since VCR can be toxic (5, 8, 9), the animals were weighed at harvest to compare body weights. There were no differences in body weights between groups, suggesting that these doses of VCR are non-toxic (Supplementary Figure S2D). Treatment with VCR reduced tumor cell proliferation and mitosis in a dose-dependent manner as demonstrated by immunostaining of RMS tumor sections for Ki-67 and PH3 (Supplementary Figures S2E, F). We next analyzed the tumor-associated angiogenesis using immunostaining for CD31, a marker of endothelial cells. There was no difference in endothelial coverage between VCR treated and control groups (Supplementary Figure S2G). The effect of VCR on apoptosis of RMS cells was not significant as shown by immunostaining for Cleaved-Caspase 3 (Supplementary Figure S2H).
Even though RCM1 has been shown previously to be a promising anti-tumor agent (19), the pre-clinical and clinical delivery of RCM1 has some limitations since RCM1 is a highly hydrophobic compound. Nanoparticles have been shown to encapsulate highly hydrophobic anti-tumor agents and are used as a vehicle for drug delivery in patients (45–49). Since nanoparticles can be administered intravenously (i.v.), we tested whether hydrophobic RCM1 compound can be delivered to RMS tumors using nanoparticles. An amphiphilic poly-beta amino ester (aPBAE) backbone was used to generate nanoparticles. The amphiphilic polymers can self-assemble with cargo in an aqueous condition to form a hydrophobic core and a hydrophilic surface, which can keep the nanoparticle stable (50, 51). Next, lecithin was added to improve nanoparticle stabilization with PEG ligands (52, 53) (Figure 3A, upper panel). Finally, the folic acid molecules were incorporated into nanoparticles to increase their specificity towards tumor cells that overexpressed folate receptor (31, 54, 55) (Figure 3A, bottom panel). Nanoparticles were labeled with DyLight 800 to visualize their recruitment into RMS tumors in mice using IVIS. Compared to nanoparticles without folic acid (NP), the nanoparticles with folic acid (NPFA) demonstrated the most efficient localization to the RMS tumors (Figure 3B), with higher average radiant efficiency (Figure 3C). Based on high tumor specificity, we used the NPFA nanoparticles thereafter. The sizes of NPFA were measured and used to calculate the hydrodynamic average diameter of NPFA as being 160.67nm (Figure 3D). The surface charge of NPFA was 38.13mV (Figure 3E), which is a suitable surface charge for tumor cell targeting (32, 56, 57). Next, RCM1 was encapsulated into NPFA nanoparticles. The UV/Vis spectrophotometry was used to determine the presence of RCM1 in the nanoparticles based on the RCM1 absorbance at 310nm and 395nm (Figure 3F). These UV/Vis spectra were used to generate a standard concentration curve and to calculate the amount of RCM1 encapsulated in the nanoparticles (Figure 3G). To test the anti-tumor efficacy of RCM1-NPFA, the tumor-bearing mice were treated with either control Empty-NPFA or RCM1-NPFA delivered i.v. every other day for 5 treatments (Figure 3H). Compared to Empty-NPFA, RCM1-containing nanoparticles reduced tumor volume in a dose-dependent manner (Figures 3I, J). A dose response curve was generated to determine the IC50 concentration for RCM1 in the nanoparticles, which was 8µg per injection (Supplementary Figure S3A). The animals were weighed at harvest. No differences in the body weights were found between mice treated with RCM1-NPFA and control Empty-NPFA (Supplementary Figure S3B). Treatment with RCM1-NPFA reduced tumor cell proliferation in a dose-dependent manner as shown by immunostaining for Ki-67 and PH3 (Supplementary Figures S3C, D). Unlike treatment with VCR in vivo, the highest dose of RCM1-NPFA reduced tumor-associated angiogenesis which was quantified as CD31+-vessel coverage within RMS tumors in mice (Supplementary Figure S3E). RCM1-NPFA treatment did not induce tumor cell apoptosis at any concentration used (Supplementary Figure S3F). Collectively, these data demonstrate that both VCR and RCM1-NPFA reduce tumor burden as single agents.
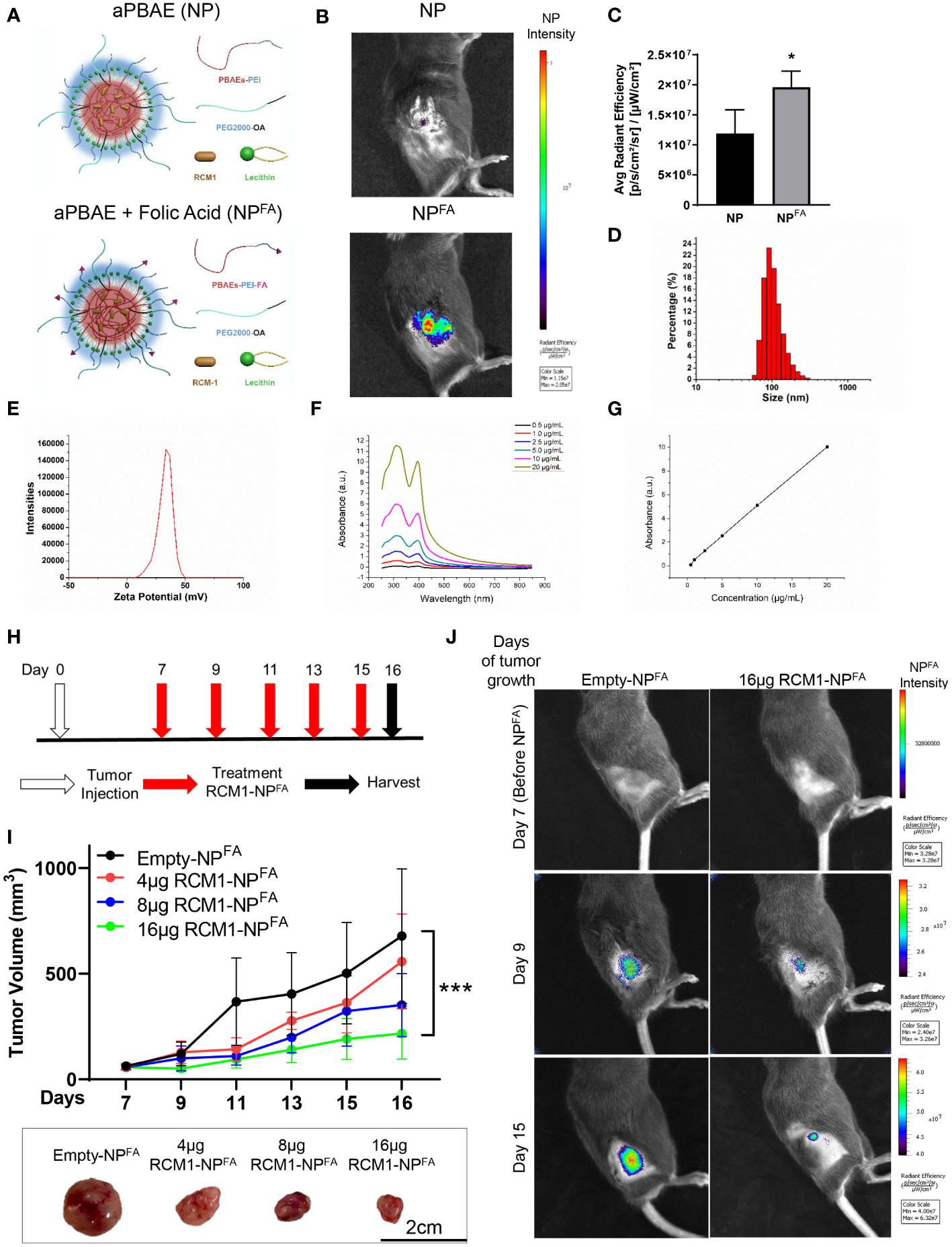
Figure 3 Generation of nanoparticle to deliver RCM1 to tumors. (A), Graphic of amphiphilic poly-beta amino ester (aPBAE) nanoparticles without folic acid (NP, top) and with folic acid (NPFA, bottom). (B), Highly efficient delivery of NPFA, compared to NP into the tumors is shown using IVIS imaging. Mice bearing Rd76-9 subcutaneous RMS tumors were injected with NP or NPFA labeled with DyLight 800. NPFA are present in the tumor 48 hours after i.v. injection. (C), Average radiant efficiency indicating NPFA have a higher intensity compared to NP. (D), The sizes of NPFA were measured and the hydrodynamic average diameter of NPFA is 160.67nm. (E), The surface charge of NPFA is 38.13mV. (F), The UV/Vis spectrum for RCM1 at increasing concentrations in DMSO determined RCM1 has an absorbance peak at 310nm and 395nm. (G), Data from the UV/VIS spectra was used to generate a standard concentration curve that was used to determine RCM1 concentration in the nanoparticles. Adjusted R2 = 0.99944. (H), Schematic diagram showing treatment strategy of tumor bearing mice. Rhabdomyosarcoma Rd76-9 cells were inoculated subcutaneously. Animals were treated with 4µg, 8µg, or 16µg RCM1- NPFA. (I), Treatment with RCM1-NPFA reduced tumor burden in a dose-dependent manner in animals. Mice were treated with 4µg, 8µg, or 16µg of RCM1-NPFA or with empty-NPFA. Tumor volumes were measured at different time points compared to empty-NPFA (top panel). Representative tumors per group are shown (bottom panel). Values are shown as mean ± SD. n=3-7, *P<0.05; ***P<0.001. (J), Presence of Empty-NPFA and RCM1-NPFA nanoparticles in the tumors are shown at days 9 and 15 after treatment. Nanoparticles were labeled with DyLight 800 and visualized using IVIS.
Combination treatment of VCR and RCM1-NPFA is more effective in reducing RMS tumor burden compared to single agents
Using the IC50 concentrations determined for both VCR (0.25mg/kg) and RCM1 (8µg/injection) in vivo (Supplementary Figure S2C and Supplementary Figure S3A respectively), we next investigated the combinatorial effects of these drugs. The Rd76-9 RMS cells were inoculated subcutaneously into the flanks of mice and allowed tumor to grow for 7 days. On day 7, the tumor-bearing mice were treated with the first dose of both VCR and RCM1. VCR was administered once a week thereafter, and RCM1-NPFA were administered every other day (Figure 4A). The combination of both drugs had shown the best efficacy in reducing tumor volumes (Figure 4B). There were no differences in body weights between control and all experimental groups of tumor-bearing mice (Supplementary Figure S4A). Blood was collected and a liver metabolic panel was analyzed from the serum of these mice. No differences were seen in the concentrations of albumin, total protein, ALP, AST, ALT, GGT and bilirubin (Supplementary Figures S4B-D). Taken together with the body weights, these results suggest that the combination therapy is non-toxic.
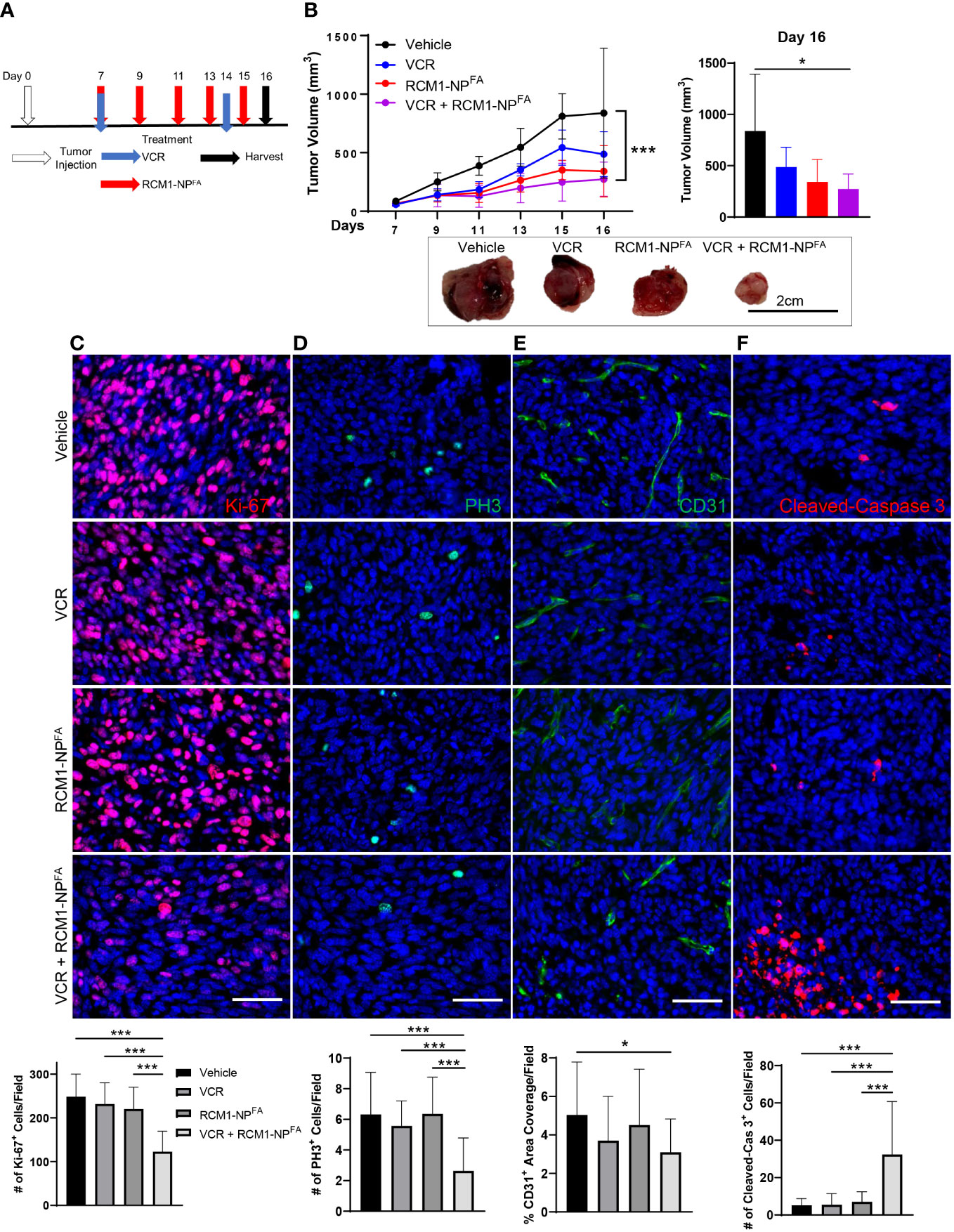
Figure 4 Combination treatment with low dose of VCR and RCM1-NPFA is more efficient than single agents in reducing rhabdomyosarcoma tumor burden in mice. (A), Experimental treatment strategy using low doses of VCR and RCM1-NPFA as single agents or in combination. Mouse rhabdomyosarcoma Rd76-9 cells were subcutaneously injected. Tumor-bearing mice were treated with IC50 dose of VCR (0.25mg/kg), IC50 dose of RCM1-NPFA (8µg) as single agents or in combination starting at day 7 after tumor cell inoculation. (B), Combination therapy reduced tumor volume compared to vehicle (saline + empty-NPFA) control. Tumor volume measurements are shown at different time points (top left) and the end of the study (day 16, top right). Representative tumors per group are shown (bottom). (C), Combination therapy is more efficient in reducing tumor cell proliferation (Ki-67, red) and mitosis (D, PH3, green) compared to single agents alone and vehicle control. (E), Combination therapy reduces tumor-associated angiogenesis (CD31, green) compared to vehicle control. (F), Combination therapy induces apoptosis (Cleaved-Caspase 3, red) compared to single agents and vehicle control. 5 random fields per sample were used to quantify the number of Ki-67+, PH3+, CD31+ and Cleaved-Caspase 3+ cells per group. Values are shown as mean ± SD. n=5-6, *P<0.05; ***P<0.001. Scale bar=50µm.
To characterize the effects of combination therapy on RMS tumors, the tumor sections were immunostained for Ki-67, PH3, CD31 and Cleaved-Caspase 3. Consistent with the in vitro data, the combination therapy synergized to reduce the number of proliferating cells in RMS tumors (Figures 4C, D). The combination therapy also reduced tumor-associated angiogenesis (Figure 4E). When assessing the cell death through Cleaved-Caspase 3 staining, we found that neither single agent at IC50 concentrations were able to increase tumor cell death. However, the combination therapy with non-toxic doses of VCR and RCM1-NPFA synergized to increase tumor cell apoptosis (Figure 4F). Our results suggest that the nanoparticle delivery of RCM1 in combination with the low doses of VCR synergizes to improve efficacy of both drugs.
The combination therapy induces a unique gene signature in RMS tumors
To determine the molecular mechanism of synergistic effect after dual treatment with RCM1-NPFA and VCR, the RNA-seq was performed to compare gene signatures of RMS tumors dissected from experimental and control groups of mice. RNA-seq analysis identified 59 differentially expressed genes in combination treated mice compared to vehicle (Figure 5A). Of which, ChaC Glutathione Specific Gamma-Glutamylcyclotransferase 1 (Chac1) was identified as one of the most downregulated genes in combination treated rhabdomyosarcoma tumors compared to vehicle-treated tumors (Figures 5A, B). Additionally, out of the 59 total differentially expressed genes in combination treated tumors, Chac1 was one of the two genes that is uniquely downregulated only in combination treated tumors, but not in either single drug treated tumors (Figure 5C). To verify the RNAseq data, we performed qRT-PCR using bulk RNA from dissected tumors. Chac1 mRNA was significantly decreased only in the tumors treated with combination therapy (Figure 5D). To further support our RNA-seq data, we examined the protein levels of CHAC1 after combination therapy in RMS tumors in vivo. Consistent with our RNA-seq and qRT-PCR data, the number of tumor cells expressing CHAC1 was reduced only after VCR+RCM1-NPFA combination treatment, but not after either single agent (Figure 5E). Using TCGA datasets, we have also demonstrated that the high expression of CHAC1 was associated with a worse prognosis in sarcoma patients (Figure 5F), suggesting that CHAC1 can play a role in RMS tumorigenesis.
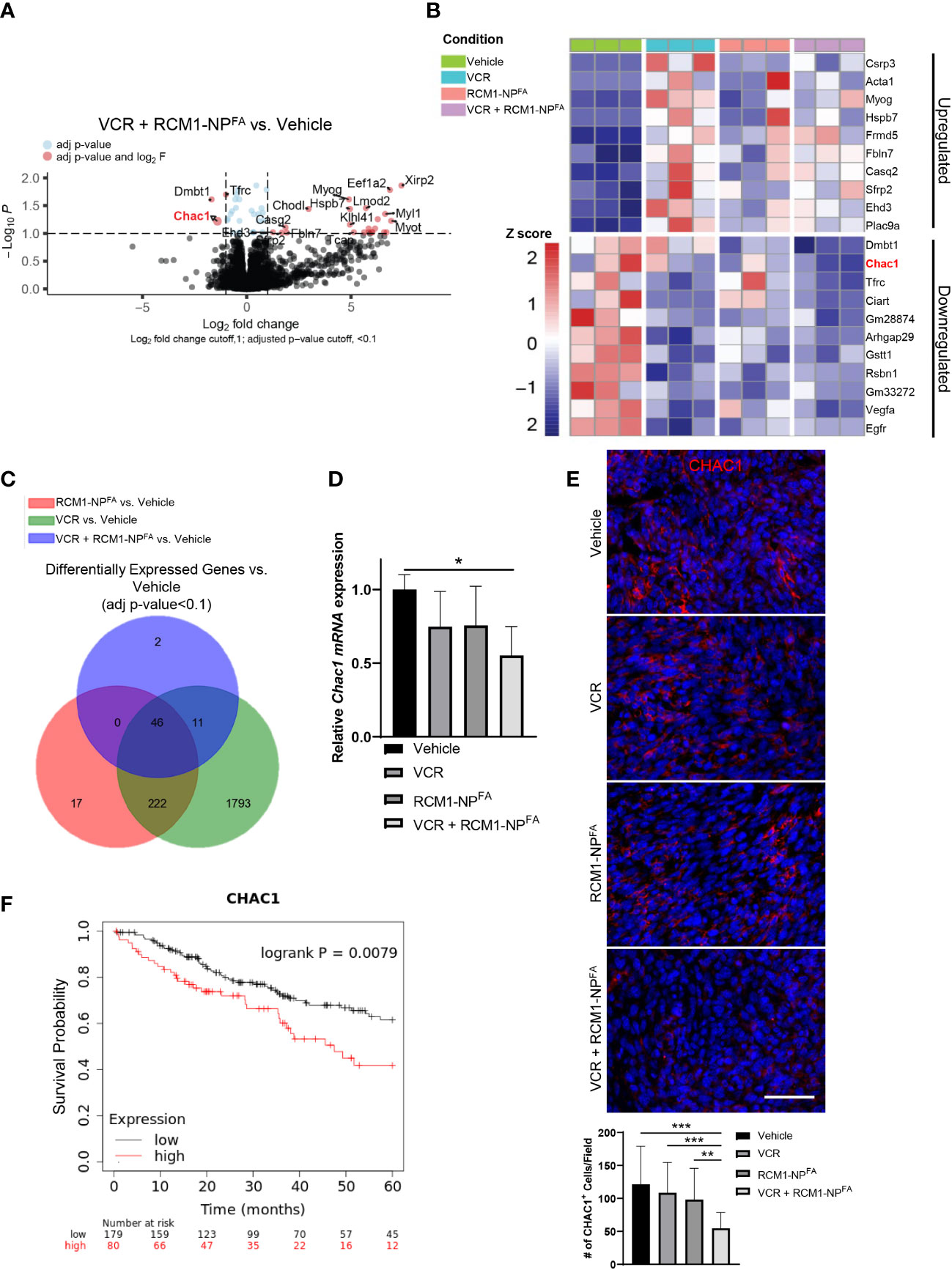
Figure 5 Treatment of mice with combination of low doses VCR and RCM1-NPFA results in unique gene signatures. (A), Volcano plot shows the differential gene expression between combo-treated (VCR + RCM1-NPFA) versus vehicle-treated (saline + empty-NPFA) tumors. RMS tumors were dissected from mice at day 16 after treatment with VCR, RCM1-NPFA alone or in combination. Genes in blue were statistically significant with an adjusted p-value <0.1. Genes in red were statistically significant with a log2 fold-change higher than 1. (B), Heat map summarizing centralized and scaled normalized counts (z-score) for the top 10 upregulated and downregulated genes in VCR + RCM1-NPFA compared to vehicle control. (C), Venn diagram showing the number of differentially expressed genes from tumors treated with VCR, RCM1-NPFA, and VCR + RCM1-NPFA compared to vehicle control. (D), Chac1 is downregulated in the combination treated group compared to vehicle control. Bars represent fold changes compared to vehicle control using beta-actin as an internal control. Values are shown as mean ± SD. n=3 per group. *, P<0.05. (E), Number of CHAC1+ cells (red) is reduced in combination therapy compared to each single agent alone and vehicle control in vivo. 5 random fields per sample were collected to quantify the number of CHAC1+ cells. Values are shown as mean ± SD. n=5-6, **P<0.01; ***P<0.001. (F), Kaplan Meyer curves demonstrating that sarcomas expressing CHAC1 above the median value (high expression) had significantly worse outcomes compared to patients with sarcomas expressing CHAC1 below the median expression value (low expression), n=259.
Knockdown of CHAC1 leads to similar effects as combination therapy
To determine the role of CHAC1 in RMS tumor cells, we generated a shRNA lentivirus against Chac1 (shChac1) and transduced Rd76-9 cells to inhibit Chac1 expression in vitro. CHAC1 protein levels in shChac1 RMS tumor cells were decreased compared to non-targeting control, confirming efficient Chac1 depletion (Figure 6A). Depletion of CHAC1 decreased the numbers of viable Rd76-9 tumor cells in culture compared to control tumor cells (Figures 6B, C). Immunostaining of Rd76-9 cells for CHAC1, Ki-67, PH3 and Cleaved-Caspase 3 protein demonstrated that the depletion of CHAC1 decreased the numbers of Ki-67+ and PH3+ proliferating tumor cells (Figures 6D-F) as well as increased the number of tumor cells undergoing apoptosis (Figure 6G). Taken together, the knockdown of CHAC1 in RMS tumor cells decreased cell proliferation and increased apoptosis, recapitulating the effects of the combination therapy with VCR and RCM1.
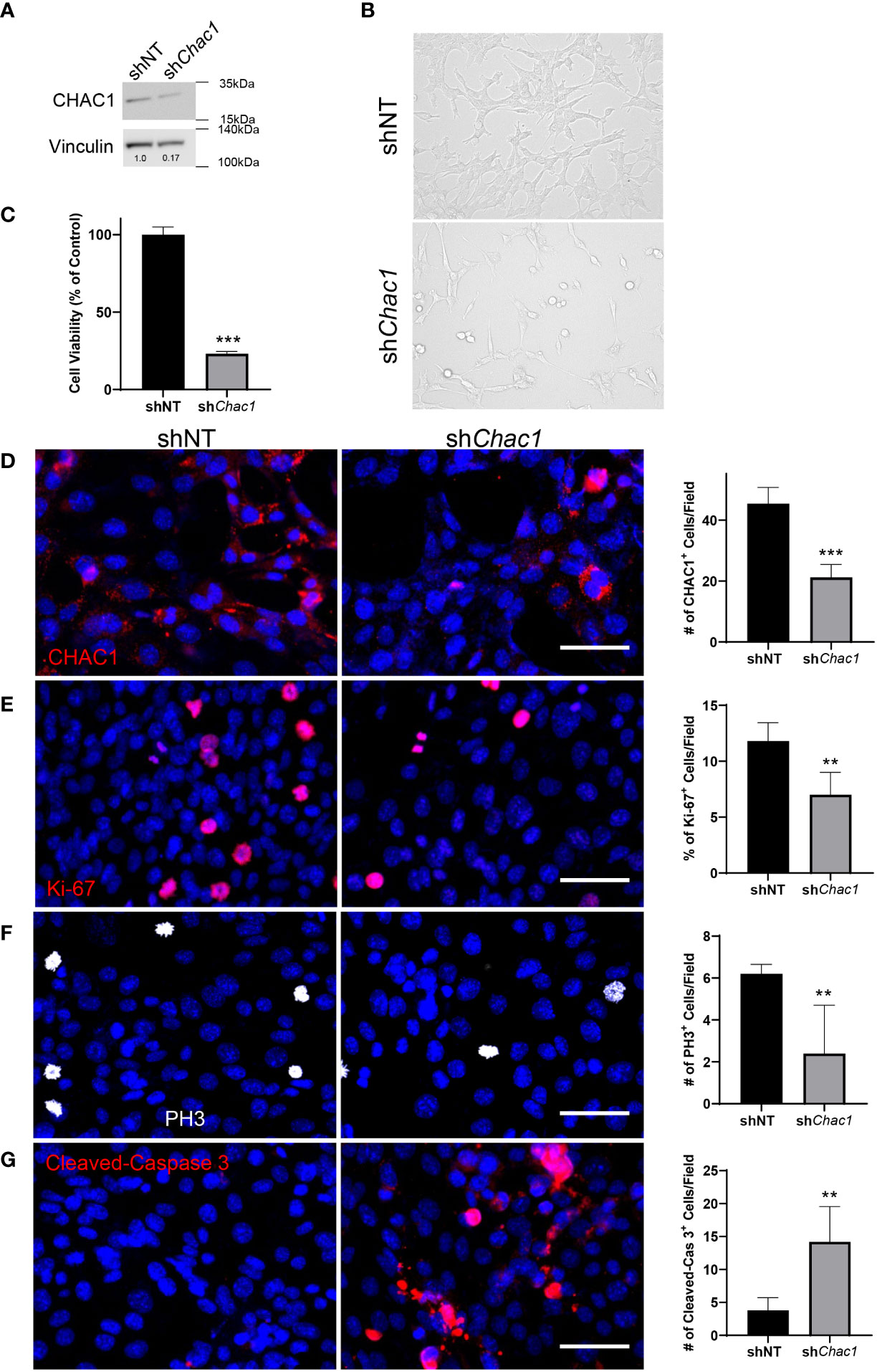
Figure 6 Depletion of Chac1 decreases rhabdomyosarcoma tumor cell growth. (A), Knockdown of Chac1 in Rd76-9 RMS cells using shChac1 efficiently decreases CHAC1 protein levels compared to shNT control, shown using Western blot. (B), Bright field images of CHAC1-deficient Rd76-9 tumor cells show increased number of floating cells and decreased number of attached cells compared to control at 24 hours after seeding. (C), Depletion of CHAC1 reduces the percent of vialble tumor cells compared to control at 48 hours after seeding. The graph shows the percentage of live cells compared to shNT control. (D), Knockdown of Chac1 in Rd76-9 RMS cells reduces the number of CHAC1+ (red) cells compared to shNT control. (E), Knockdown of Chac1 in Rd76-9 RMS cells reduces proliferation (Ki-67, red) and mitosis (F, PH3, white) compared to shNT. (G), Knockdown of Chac1 induces apoptosis in Rd76-9 tumor cells (Cleaved-Caspase 3, red) compared to shNT control. 5 random fields per sample were used to quantify the numbers of CHAC1+, Ki-67+, PH3+ and Cleaved-Caspase 3+ cells per group. Values are shown as mean ± SD. **P<0.01; ***P<0.001. Scale bar=50µm.
Discussion
Rhabdomyosarcoma (RMS) is a highly aggressive pediatric soft tissue cancer with a poor prognosis (2–4). Vincristine (VCR) is a cell-cycle specific chemotherapeutic that is a first line of defense for these patients (7). However, VCR can be toxic (5), and many patients relapse or develop therapeutic resistance (2, 3), demonstrating an urgent need for new treatment strategies.
FOXM1 is an oncogene that is overexpressed in many cancers and has been clinically associated with a worse prognosis (15, 19). Despite the well-known roles of FOXM1 in cancer, there has been a lack of investigation of FOXM1 in RMS. FOXM1 is primarily expressed during embryogenesis, with minimal-to-no post-natal expression in normal healthy tissue, making it a very attractive therapeutic target (18, 19). There are many reports of pharmacological inhibition of FOXM1 (23, 24, 58). However, many of these drugs lack specificity, creating off-target effects, side effects, and are toxic (17, 23, 58). To date, there are not any clinically approved drugs to target FOXM1. We recently identified a small molecule inhibitor of FOXM1, Robert Costa Memorial Drug-1 (RCM1) that decreased tumorigenesis across multiple cancer types, including RMS, and is also non-toxic (19). FOXM1 is important in the cell-cycle, and interestingly, treatment with RCM1 increases the duration of the cell-cycle, suggesting that RCM1 can be used with other cell-cycle specific anticancer therapies for combinatorial effects.
In the present study, we demonstrated that low-dose concentrations of VCR and RCM1 in combination synergized to reduce tumor cells proliferation and mitosis across mouse and human RMS cell lines compared to each single agent. Remarkably, while neither low-dose concentration of VCR nor RCM1 could induce apoptosis alone, the combination therapy synergized to induce apoptosis of tumor cells in vitro.
To maintain clinical relevance, we generated VCR dose response curves in vivo using doses near the animal equivalency dose for patients (59). In addition, we administered VCR every 7 days, consistent with clinical use. As expected, the tumor burden and proliferation were reduced in a dose-dependent manner. Interestingly, these doses of VCR did not induce apoptosis, which could be explained due to the fact that the dosage used was low to avoid toxicity.
While RCM1 has been shown to be a promising anticancer therapy (19), there has been limitations to administration due to the high hydrophobicity of the drug. Previous reports have only been able to dissolve RCM1 in either pure DMSO, or a DMSO mixed solution, which is not a clinically relevant vehicle (19, 25). To make RCM1 administration more clinically relevant, we synthesized amphiphilic poly-beta amino ester (aPBAE) nanoparticles to use as a vehicle for RCM1 delivery. Nanoparticles have been used as a vehicle in the clinic for hydrophobic drugs like paclitaxel, as well as other RMS therapeutics like doxorubicin, and can be administered intravenously for several cancer types (60, 61). The aPBAE is one of the polymers that is used in generating nanoparticles and has recently attracted enormous attention in gene or drug delivery due to its excellent biocompatibility and biodegradability (62). The published studies combined aPBAEs and lipid molecules, including lecithin, to improve stability of nanoparticles (63). To further improve nanoparticle stabilization, lecithin was combined with PEG ligands, especially in liposomes or polymer-lipid hybrid nanoparticles (52, 53). To help guide the nanoparticles to the tumor, the newly synthesized aPBAE nanoparticles are also combined with folic acid, a metabolite that is readily up taken by tumors, due to the over-expression of folate acid receptors on the surfaces of tumor cells (31, 64). We demonstrated that RCM1-encapsulated nanoparticles can be injected intravenously and localize at the site of the tumor. Considering FOXM1 has minimal-to-no expression in normal healthy tissue, we did not expect any off-target effects. Mice treated with RCM1-encapsulated nanoparticles did not show any differences in body weights, suggesting RCM1-encapsulated nanoparticles are non-toxic, which is consistent with previous studies of RCM1 (19). The RCM1-encapsulated nanoparticles reduce tumor burden, proliferation, mitosis, and angiogenesis in a dose-dependent manner.
While both VCR and RCM1-encapsulated nanoparticles were able to reduce tumor burden in a dose-dependent manner, we selected IC50 concentrations based off of our dose response curves to use in combination in vivo. Impressively, while neither IC50 concentration of VCR nor RCM1 could reduce tumor burden, the combination therapy with the same doses reduced tumor burden. Consistent with our in vitro findings, the combination therapy synergized to reduce proliferation and mitosis, while also inducing apoptosis. The IC50 concentrations of VCR and RCM1 as single agents did not impact angiogenesis, however, the combination therapy was able to reduce angiogenesis. Altogether, our data suggests that the combination therapy is superior to VCR and RCM1 as single agents.
To determine the mechanism of this novel combination therapy, we performed bulk RNA-seq. We identified Chac1 as a uniquely downregulated gene in the combination treated group compared to singles agents and control. Chac1 is a gene implicated in regulation of cell death, ER stress and glutathione biosynthesis (65, 66), but the role of Chac1 in RMS has not been studied. Interestingly, high levels of CHAC1 are associated with a worse prognosis in patients (16, 67). Previous reports have shown that anti-tumor agents can reduce CHAC1 levels in cancer (68, 69). However, there have been no reports on the role of CHAC1 in RMS. Lentiviral knockdown of Chac1 in murine RMS cells caused a reduction of proliferation, mitosis, and induction of apoptosis. These phenotypes are similar to VCR and RCM1 combination therapy in vitro and in vivo.
While this study provides insight for a novel combination therapy for RMS, there are limitations. All in vivo experiments are conducted within 16 days, due to the untreated tumors rapid growth that required euthanasia per our IACUC protocol. This is a relatively short time and does not allow prolonged examination of the effects of the combination therapy. As a future direction, a survival study could be performed where each animal will be treated until euthanasia criteria are met, including tumor volumes, body weights, etc. Also, the current study has not assessed whether the RCM1/VCR combination therapy could be universally effective for the different subtypes of RMS, including embryonal, alveolar, pleomorphic, and spindle cell/sclerosing variants. Since it is well known that all these factors could greatly affect the chemosensitivity of the tumor (70). Additionally, all in vivo studies have been conducted using mouse models. While this is a widely accepted condition, it may not recapitulate the possible effects observed in patients. It is unclear whether nanoparticle delivery of RCM1/VCR combination therapy will have beneficial effect in human RMS. The efficacy of RCM1/VCR combination therapy in RMS can only be determined in clinical trials.
Data availability statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession number GSE223149 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223149).
Ethics statement
All animal studies were approved by Cincinnati Children’s Research Foundation Institutional Animal Care and Use Committee and covered under our animal protocol (IACUC2020-0041).
Author contributions
Conception and Design: JD, TK. Development of methodology: JD, ZD, SS, FB, JG-A. Acquisition of data: JD, ZD, FB, SS, JG-A. Analysis and interpretation of data: JD, ZD, FB, SS, JG-A, TK. Writing, review and/or revision of the manuscript: JD, ZD, FB, SS, JG-A, DS, VK, TK. Study Supervison: TK. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Tien Le for technical assistance. This work was supported by the NIH grants R01 HL132849 (TK), R01 HL158659 (TK), R01 HL141174 (VK), R01 HL149631 (VK), R01 HL152973 (VK/TK), as well as the Graduate Student Government Research Fellowship at the University of Cincinnati (JD).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1112859/full#supplementary-material
Supplementary Figure 1 | Bright field images showing that combination treatment with low doses of vincristine and RCM1 is more efficient in decreasing growth of mouse and human rhabdomyosarcoma cells compared to single agents. (A) Combination of IC50 dose for VCR (2.1nm) and RCM1 (8.7µM) reduced the number of tumor cells compared to IC50 doses of each single agent alone and vehicle (saline + DMSO) control in murine RMS in vitro. (B) Combination of IC50 concentrations for VCR (3.0nm) and RCM1 (3.5µM) reduced the number of tumor cells compared to IC50 doses of each single agent alone and vehicle (saline + DMSO) control in human RMS in vitro. Scale bar=50µm.
Supplementary Figure 2 | Treatment with vincristine reduces tumor burden in a dose-dependent manner in a mouse models of rhabdomyosarcoma. (A) Experimental VCR treatment strategy using Rd76-9 as a subcutaneous tumor model. Animals were treated with 0.1mg/kg, 0.2mg/kg or 0.4mg/kg VCR. (B) VCR reduced tumor volume compared to vehicle (saline) control. Tumor volume measurements are shown at different time-points (top) and representative tumor images per group (bottom). (C) The dose-response curve, showing tumor volumes at day 16 after treatment with different doses of VCR, was used to determine the IC50 concentration of VCR in vivo (0.25mg/kg). (D) No differences were found in body weights of mice from each group. (E) VCR reduces proliferation (Ki-67, red) and mitosis ((F) PH3, green) of tumor cells compared to vehicle control. (G) VCR does not reduce tumor-associated angiogenesis (CD31, green) compared to vehicle control. (H) VCR does not induce apoptosis (Cleaved-Caspase 3, red) compared vehicle control. 5 random fields per sample were collected to quantify the number of Ki-67+, PH3+, CD31+ and Cleaved-Caspase 3+ cells per group. Values are shown as mean ± SD. n=4, *, P<0.05; ***, P<0.001. Scale bar=50µm.
Supplementary Figure 3 | Treatment with RCM1-NPFA reduces tumor growth in a dose-dependent manner in a mouse models of rhabdomyosarcoma. (A) The dose-response curve, showing tumor volumes at day 16 after treatment with different doses of RCM1-NPFA, was used to determine the IC50 concentration of RCM1-NPFA in vivo (8µg). (B) No differences in body weights of mice from each group were found. (C) RCM1-NPFA reduced tumor cell proliferation (Ki-67, red) and mitosis (D PH3, green) compared to vehicle (empty-NPFA) control. (E) RCM1-NPFA reduces tumor-associated angiogensis (CD31, green) compared to vehicle control. (F) RCM1-NPFA does not induce apoptosis of tumor cells (Cleaved-Caspase 3, red) compared vehicle control. 5 random fields per sample were collected to quantify the number of Ki-67+, PH3+, CD31+ and Cleaved-Caspase 3+ cells per group. Values are shown as mean ± SD. n=3-7, **, P<0.01; ***, P<0.001. Scale bar=50µm.
Supplementary Figure 4 | Combination therapy with low doses of VCR and RCM1-NPFA is non-toxic in a mouse model of rhabdomyosarcoma. (A) No differences were found in body weights of mice from different groups treated either with single agents or with combination of VCR and RCM1-NPFA. (B–D) No differences were found between groups across different liver enzymes in serum. Saline + empty-NPFA was used as a vehicle control. Values are shown as mean ± SD. n=5-6 per group, P>0.05.
References
1. Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the united states, 1975-2005. Cancer (2009) 115(18):4218–26. doi: 10.1002/cncr.24465
2. Xu N, Hua Z, Ba G, Zhang S, Liu Z, Thiele CJ, et al. The anti-tumor growth effect of a novel agent DMAMCL in rhabdomyosarcoma in vitro and in vivo. J Exp Clin Cancer Res: CR (2019) 38(1):118. doi: 10.1186/s13046-019-1107-1
3. Bharathy N, Berlow NE, Wang E, Abraham J, Settelmeyer TP, Hooper JE, et al. The HDAC3-SMARCA4-miR-27a axis promotes expression of the PAX3:FOXO1 fusion oncogene in rhabdomyosarcoma. Sci Signal (2018) 11(557). doi: 10.1126/scisignal.aau7632
4. Miyoshi K, Kohashi K, Fushimi F, Yamamoto H, Kishimoto J, Taguchi T, et al. Close correlation between CXCR4 and VEGF expression and frequent CXCR7 expression in rhabdomyosarcoma. Hum Pathol (2014) 45(9):1900–9. doi: 10.1016/j.humpath.2014.05.012
5. Mu Y, Liu Y, Li L, Tian C, Zhou H, Zhang Q, et al. The novel tubulin polymerization inhibitor MHPT exhibits selective anti-tumor activity against rhabdomyosarcoma in vitro and in vivo. PloS One (2015) 10(3):e0121806. doi: 10.1371/journal.pone.0121806
6. De Vita A, Vanni S, Fausti V, Cocchi C, Recine F, Miserocchi G, et al. Deciphering the genomic landscape and pharmacological profile of uncommon entities of adult rhabdomyosarcomas. Int J Mol Sci (2021) 22(21). doi: 10.3390/ijms222111564
7. Kurmasheva RT, Bandyopadhyay A, Favours E, Del Pozo V, Ghilu S, Phelps DA, et al. Evaluation of entinostat alone and in combination with standard-of-care cytotoxic agents against rhabdomyosarcoma xenograft models. Pediatr Blood Cancer (2019) 66(8):e27820. doi: 10.1002/pbc.27820
8. Lee JY, Sim WS, Cho NR, Kim BW, Moon JY, Park HJ. The antiallodynic effect of nefopam on vincristine-induced neuropathy in mice. J Pain Res (2020) 13:323–29. doi: 10.2147/JPR.S224478
9. Eberle-Singh JA, Sagalovskiy I, Maurer HC, Sastra SA, Palermo CF, Decker AR, et al. Effective delivery of a microtubule polymerization inhibitor synergizes with standard regimens in models of pancreatic ductal adenocarcinoma. Clin Cancer Res (2019) 25(18):5548–60. doi: 10.1158/1078-0432.CCR-18-3281
10. Balli D, Zhang Y, Snyder J, Kalinichenko VV, Kalin TV. Endothelial cell-specific deletion of transcription factor FoxM1 increases urethane-induced lung carcinogenesis. Cancer Res (2011) 71(1):40–50. doi: 10.1158/0008-5472.CAN-10-2004
11. Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle (2011) 10(3):396–405. doi: 10.4161/cc.10.3.14709
12. Balli D, Ren X, Chou FS, Cross E, Zhang Y, Kalinichenko VV, et al. Foxm1 transcription factor is required for macrophage migration during lung inflammation and tumor formation. Oncogene (2012) 31(34):3875–88. doi: 10.1038/onc.2011.549
13. Milewski D, Balli D, Ustiyan V, Le T, Dienemann H, Warth A, et al. FOXM1 activates AGR2 and causes progression of lung adenomas into invasive mucinous adenocarcinomas. PloS Genet (2017) 13(12):e1007097. doi: 10.1371/journal.pgen.1007097
14. Wang IC, Meliton L, Tretiakova M, Costa RH, Kalinichenko VV, Kalin TV. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene (2008) 27(30):4137–49. doi: 10.1038/onc.2008.60
15. Kuda M, Kohashi K, Yamada Y, Maekawa A, Kinoshita Y, Nakatsura T, et al. FOXM1 expression in rhabdomyosarcoma: A novel prognostic factor and therapeutic target. Tumour Biol (2016) 37(4):5213–23. doi: 10.1007/s13277-015-4351-9
16. Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep (2021) 11(1):6047. doi: 10.1038/s41598-021-84787-5
17. Kalinichenko VV, Kalin TV. Is there potential to target FOXM1 for ‘undruggable’ lung cancers? Expert Opin Ther Targets (2015) 19(7):865–7. doi: 10.1517/14728222.2015.1042366
18. Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: Repurposing an oncogene as a biomarker. Semin Cancer Biol (2018) 52(Pt 1):74–84. doi: 10.1016/j.semcancer.2017.08.009
19. Shukla S, Milewski D, Pradhan A, Rama N, Rice K, Le T, et al. The FOXM1 inhibitor RCM-1 decreases carcinogenesis and nuclear beta-catenin. Mol Cancer Ther (2019) 18(7):1217–29. doi: 10.1158/1535-7163.MCT-18-0709
20. Black M, Arumugam P, Shukla S, Pradhan A, Ustiyan V, Milewski D, et al. FOXM1 nuclear transcription factor translocates into mitochondria and inhibits oxidative phosphorylation. Mol Biol Cell (2020) 31(13):1411–24. doi: 10.1091/mbc.E19-07-0413
21. Wang IC, Meliton L, Ren X, Zhang Y, Balli D, Snyder J, et al. Deletion of forkhead box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PloS One (2009) 4(8):e6609. doi: 10.1371/journal.pone.0006609
22. Cai Y, Balli D, Ustiyan V, Fulford L, Hiller A, Misetic V, et al. Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. J Biol Chem (2013) 288(31):22527–41. doi: 10.1074/jbc.M113.455089
23. Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther (2008) 7(7):2022–32. doi: 10.1158/1535-7163.MCT-08-0188
24. Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta (2012) 1819(1):28–37. doi: 10.1016/j.bbagrm.2011.09.004
25. Sun L, Ren X, Wang IC, Pradhan A, Zhang Y, Flood HM, et al. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci Signal (2017) 10(475). doi: 10.1126/scisignal.aai8583
26. McAllister RM, Melnyk J, Finkelstein JZ, Adams EC Jr., Gardner MB. Cultivation in vitro of cells derived from a human rhabdomyosarcoma. Cancer (1969) 24(3):520–6. doi: 10.1002/1097-0142(196909)24:3<520::AID-CNCR2820240313>3.0.CO;2-M
27. Missiaglia E, Selfe J, Hamdi M, Williamson D, Schaaf G, Fang C, et al. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: An approach to identify candidate genes involved in tumor development. Genes Chromosomes Cancer (2009) 48(6):455–67. doi: 10.1002/gcc.20655
28. Schaaf G, Hamdi M, Zwijnenburg D, Lakeman A, Geerts D, Versteeg R, et al. Silencing of SPRY1 triggers complete regression of rhabdomyosarcoma tumors carrying a mutated RAS gene. Cancer Res (2010) 70(2):762–71. doi: 10.1158/0008-5472.CAN-09-2532
29. Houghton JA, Houghton PJ, Green AA. Chemotherapy of childhood rhabdomyosarcomas growing as xenografts in immune-deprived mice. Cancer Res (1982) 42(2):535–9.
30. Leddon JL, Chen CY, Currier MA, Wang PY, Jung FA, Denton NL, et al. Oncolytic HSV virotherapy in murine sarcomas differentially triggers an antitumor T-cell response in the absence of virus permissivity. Mol Ther Oncolytics (2015) 1:14010. doi: 10.1038/mto.2014.10
31. Deng Z, Lin J, Bud’ko SL, Webster B, Kalin TV, Kalinichenko VV, et al. Dual targeting with cell surface electrical charge and folic acid via superparamagnetic Fe(3)O(4)@Cu(2-x)S for photothermal cancer cell killing. Cancers (2021) 13(21). doi: 10.3390/cancers13215275
32. Dunn AW, Kalinichenko VV, Shi D. Highly efficient In vivo targeting of the pulmonary endothelium using novel modifications of polyethylenimine: An importance of charge. Adv Healthc Mater (2018) 7(23):e1800876. doi: 10.1002/adhm.201800876
33. Hu C, Zhang F, Long L, Kong Q, Luo R, Wang Y. Dual-responsive injectable hydrogels encapsulating drug-loaded micelles for on-demand antimicrobial activity and accelerated wound healing. J Control Release (2020) 324:204–17. doi: 10.1016/j.jconrel.2020.05.010
34. Liu Y, Fan Q, Huo Y, Liu C, Li B, Li Y. Construction of a mesoporous Polydopamine@GO/Cellulose nanofibril composite hydrogel with an encapsulation structure for controllable drug release and toxicity shielding. ACS Appl Materials Interfaces (2020) 12(51):57410–20. doi: 10.1021/acsami.0c15465
35. Coletta A, Castelli S, Chillemi G, Sanna N, Cushman M, Pommier Y, et al. Solvent dependency of the UV-vis spectrum of indenoisoquinolines: Role of keto-oxygens as polarity interaction probes. PloS One (2013) 8(9):e73881. doi: 10.1371/journal.pone.0073881
36. Black M, Milewski D, Le T, Ren X, Xu Y, Kalinichenko VV, et al. FOXF1 inhibits pulmonary fibrosis by preventing CDH2-CDH11 cadherin switch in myofibroblasts. Cell Rep (2018) 23(2):442–58. doi: 10.1016/j.celrep.2018.03.067
37. Cheng XH, Black M, Ustiyan V, Le T, Fulford L, Sridharan A, et al. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PloS Genet (2014) 10(9):e1004656. doi: 10.1371/journal.pgen.1004656
38. Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am J Respir Cell Mol Biol (2008) 39(4):390–9. doi: 10.1165/rcmb.2008-0044OC
39. Milewski D, Shukla S, Gryder BE, Pradhan A, Donovan J, Sudha P, et al. FOXF1 is required for the oncogenic properties of PAX3-FOXO1 in rhabdomyosarcoma. Oncogene (2021) 40(12):2182–99. doi: 10.1038/s41388-021-01694-9
40. Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B, et al. FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21(Cip1) CDK inhibitor. Oncogene (2017) 36(6):850–62. doi: 10.1038/onc.2016.254
41. Bolte C, Ren X, Tomley T, Ustiyan V, Pradhan A, Hoggatt A, et al. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J Biol Chem (2015) 290(12):7563–75. doi: 10.1074/jbc.M114.609487
42. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol (2016) 34(5):525–7. doi: 10.1038/nbt.3519
43. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol (2014) 15(12):550. doi: 10.1186/s13059-014-0550-8
44. Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res (2010) 38(Web Server issue):W755–62. doi: 10.1093/nar/gkq405
45. Wang Y, Sun S, Zhang Z, Shi D. Nanomaterials for cancer precision medicine. Adv Mater (2018) 30(17):e1705660. doi: 10.1002/adma.201705660
46. Xu J, Cheng X, Tan L, Fu C, Ahmed M, Tian J, et al. Microwave responsive nanoplatform via p-selectin mediated drug delivery for treatment of hepatocellular carcinoma with distant metastasis. Nano Lett (2019) 19(5):2914–27. doi: 10.1021/acs.nanolett.8b05202
47. Deng Z, Kalin GT, Shi D, Kalinichenko VV. Nanoparticle delivery systems with cell-specific targeting for pulmonary diseases. Am J Respir Cell Mol Biol (2021) 64(3):292–307. doi: 10.1165/rcmb.2020-0306TR
48. Gagliardi A, Giuliano E, Venkateswararao E, Fresta M, Bulotta S, Awasthi V, et al. Biodegradable polymeric nanoparticles for drug delivery to solid tumors. Front Pharmacol (2021) 12:601626. doi: 10.3389/fphar.2021.601626
49. Sun S, Wang Y, Zhou R, Deng Z, Han Y, Han X, et al. Targeting and regulating of an oncogene via nanovector delivery of MicroRNA using patient-derived xenografts. Theranostics (2017) 7(3):677–93. doi: 10.7150/thno.16357
50. Zhang L, Chan JM, Gu FX, Rhee JW, Wang AZ, Radovic-Moreno AF, et al. Self-assembled lipid–polymer hybrid nanoparticles: a robust drug delivery platform. ACS Nano (2008) 2(8):1696–702. doi: 10.1021/nn800275r
51. Zhang LaZ L. LIPID–POLYMER HYBRID NANOPARTICLES: SYNTHESIS, CHARACTERIZATION AND APPLICATIONS. Nano Life (2010) 01(01n02):163–73. doi: 10.1142/S179398441000016X
52. Le NTT, Cao VD, Nguyen TNQ, Le TTH, Tran TT, Hoang Thi TT. Soy lecithin-derived liposomal delivery systems: Surface modification and current applications. Int J Mol Sci (2019) 20(19). doi: 10.3390/ijms20194706
53. Zhu C, Huo D, Chen Q, Xue J, Shen S, Xia Y. A eutectic mixture of natural fatty acids can serve as the gating material for near-Infrared-Triggered drug release. Adv Mater (2017) 29(40). doi: 10.1002/adma.201703702
54. Yu J, He X, Wang Z, Wang Y, Liu S, Li X, et al. Combining PD-L1 inhibitors with immunogenic cell death triggered by chemo-photothermal therapy via a thermosensitive liposome system to stimulate tumor-specific immunological response. Nanoscale (2021) 13(30):12966–78. doi: 10.1039/D1NR03288G
55. Sun C, Zhang H, Li S, Zhang X, Cheng Q, Ding Y, et al. Polymeric nanomedicine with “Lego” surface allowing modular functionalization and drug encapsulation. ACS Appl Materials Interfaces (2018) 10(30):25090–98. doi: 10.1021/acsami.8b06598
56. Chen B, Le W, Wang Y, Li Z, Wang D, Ren L, et al. Targeting negative surface charges of cancer cells by multifunctional nanoprobes. Theranostics (2016) 6(11):1887–98. doi: 10.7150/thno.16358
57. Han X, Deng Z, Yang Z, Wang Y, Zhu H, Chen B, et al. Biomarkerless targeting and photothermal cancer cell killing by surface-electrically-charged superparamagnetic Fe(3)O(4) composite nanoparticles. Nanoscale (2017) 9(4):1457–65. doi: 10.1039/C6NR07161A
58. Gartel AL. A new target for proteasome inhibitors: FoxM1. Expert Opin Investig Drugs (2010) 19(2):235–42. doi: 10.1517/13543780903563364
59. Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm (2016) 7(2):27–31. doi: 10.4103/0976-0105.177703
60. Ganugula R, Deng M, Arora M, Pan HL, Kumar M. Polyester nanoparticle encapsulation mitigates paclitaxel-induced peripheral neuropathy. ACS Chem Neurosci (2019) 10(3):1801–12. doi: 10.1021/acschemneuro.8b00703
61. Mercatali L, Vanni S, Miserocchi G, Liverani C, Spadazzi C, Cocchi C, et al. The emerging role of cancer nanotechnology in the panorama of sarcoma. Front Bioeng Biotechnol (2022) 10:953555. doi: 10.3389/fbioe.2022.953555
62. Liu Y, Li Y, Keskin D, Shi L. Poly(beta-amino esters): Synthesis, formulations, and their biomedical applications. Adv Healthc Mater (2019) 8(2):e1801359. doi: 10.1002/adhm.201801359
63. Kaczmarek JC, Kauffman KJ, Fenton OS, Sadtler K, Patel AK, Heartlein MW, et al. Optimization of a degradable polymer-lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett (2018) 18(10):6449–54. doi: 10.1021/acs.nanolett.8b02917
64. Hanley KZ, Horowitz IR, Gordon A, Meisel J, Khanna N. Folate receptor alpha is preferentially expressed in the carcinoma component of endometrial carcinosarcomas: A potential target for adjuvant therapy. Int J Gynecological Pathol (2021) 40(5):501–09. doi: 10.1097/PGP.0000000000000736
65. Kumar A, Tikoo S, Maity S, Sengupta S, Sengupta S, Kaur A, et al. Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep (2012) 13(12):1095–101. doi: 10.1038/embor.2012.156
66. Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife (2014) 3:e02523. doi: 10.7554/eLife.02523
67. Lanczky A, Gyorffy B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J Med Internet Res (2021) 23(7):e27633. doi: 10.2196/27633
68. Sertorio M, Nowrouzi A, Akbarpour M, ChEtal K, Salomonis N, Brons S, et al. Differential transcriptome response to proton versus X-ray radiation reveals novel candidate targets for combinatorial PT therapy in lymphoma. Radiother Oncol (2021) 155:293–303. doi: 10.1016/j.radonc.2020.10.024
69. Kim DH, Sun D, Storck WK, Welker Leng K, Jenkins C, Coleman DJ, et al. BET bromodomain inhibition blocks an AR-repressed, E2F1-activated treatment-emergent neuroendocrine prostate cancer lineage plasticity program. Clin Cancer Res (2021) 27(17):4923–36. doi: 10.1158/1078-0432.CCR-20-4968
Keywords: FOXM1 inhibitor, combination therapy, rhabdomyosarcoma, nanoparticles, animal models
Citation: Donovan J, Deng Z, Bian F, Shukla S, Gomez-Arroyo J, Shi D, Kalinichenko VV and Kalin TV (2023) Improving anti-tumor efficacy of low-dose Vincristine in rhabdomyosarcoma via the combination therapy with FOXM1 inhibitor RCM1. Front. Oncol. 13:1112859. doi: 10.3389/fonc.2023.1112859
Received: 30 November 2022; Accepted: 06 January 2023;
Published: 02 February 2023.
Edited by:
Keqiang Zhang, City of Hope National Medical Center, United StatesCopyright © 2023 Donovan, Deng, Bian, Shukla, Gomez-Arroyo, Shi, Kalinichenko and Kalin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tanya V. Kalin, tatiana.kalin@cchmc.org
†These authors have contributed equally to this work
 Johnny Donovan
Johnny Donovan Zicheng Deng1,2,3†
Zicheng Deng1,2,3† Samriddhi Shukla
Samriddhi Shukla Tanya V. Kalin
Tanya V. Kalin