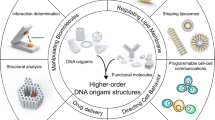
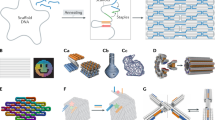
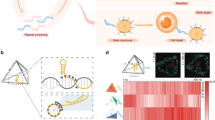
Abstract
DNA origami has emerged as a powerful method to generate DNA nanostructures with dynamic properties and nanoscale control. These nanostructures enable complex biophysical studies and the fabrication of next-generation therapeutic devices. For these applications, DNA origami typically needs to be functionalized with bioactive ligands and biomacromolecular cargos. Here, we review methods developed to functionalize, purify and characterize DNA origami nanostructures. We identify remaining challenges such as limitations in functionalization efficiency and characterization. We then discuss where researchers can contribute to further advance the fabrication of functionalized DNA origami.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hong, F., Zhang, F., Liu, Y. & Yan, H. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 117, 12584–12640 (2017).
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Ke, Y., Ong, L. L., Shih, W. M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012).
Benson, E. et al. DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015).
Veneziano, R. et al. Designer nanoscale DNA assemblies programmed from the top down. Science 352, 1534 (2016).
Tikhomirov, G., Petersen, P. & Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 552, 67–71 (2017).
Wagenbauer, K. F., Sigl, C. & Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 552, 78–83 (2017).
Yao, G. et al. Meta-DNA structures. Nat. Chem. 12, 1067–1075 (2020).
Huang, C. M., Kucinic, A., Johnson, J. A., Su, H. J. & Castro, C. E. Integrated computer-aided engineering and design for DNA assemblies. Nat. Mater. 20, 1264–1271 (2021).
Jun, H. et al. Autonomously designed free-form 2D DNA origami. Sci. Adv. 5, eaav0655 (2019).
Jun, H. et al. Automated sequence design of 3D polyhedral wireframe DNA origami with honeycomb edges. ACS Nano 13, 2083–2093 (2019).
Jun, H., Wang, X., Bricker, W. P. & Bathe, M. Automated sequence design of 2D wireframe DNA origami with honeycomb edges. Nat. Commun. 10, 5419 (2019).
Jun, H. et al. Rapid prototyping of arbitrary 2D and 3D wireframe DNA origami. Nucleic Acids Res. 49, 10265–10274 (2021).
Veneziano, R. et al. In vitro synthesis of gene-length single-stranded DNA. Sci. Rep. 8, 6548 (2018).
Nafisi, P. M., Aksel, T. & Douglas, S. M. Construction of a novel phagemid to produce custom DNA origami scaffolds. Synth. Biol. 3, ysy015 (2018).
Shepherd, T. R., Du, R. R., Huang, H., Wamhoff, E. C. & Bathe, M. Bioproduction of pure, kilobase-scale single-stranded DNA. Sci. Rep. 9, 6121 (2019).
Engelhardt, F. A. S. et al. Custom-size, functional, and durable DNA origami with design-specific scaffolds. ACS Nano 13, 5015–5027 (2019).
Minev, D. et al. Rapid in vitro production of single-stranded DNA. Nucleic Acids Res. 47, 11956–11962 (2019).
Noteborn, W. E. M., Abendstein, L. & Sharp, T. H. One-pot synthesis of defined-length ssDNA for multiscaffold DNA origami. Bioconjug. Chem. 32, 94–98 (2021).
Praetorius, F. et al. Biotechnological mass production of DNA origami. Nature 552, 84–87 (2017).
Ducani, C., Kaul, C., Moche, M., Shih, W. M. & Hogberg, B. Enzymatic production of ‘monoclonal stoichiometric’ single-stranded DNA oligonucleotides. Nat. Methods 10, 647–652 (2013).
Schmidt, T. L. et al. Scalable amplification of strand subsets from chip-synthesized oligonucleotide libraries. Nat. Commun. 6, 8634 (2015).
Halley, P. D., Patton, R. A., Chowdhury, A., Byrd, J. C. & Castro, C. E. Low-cost, simple, and scalable self-assembly of DNA origami nanostructures. Nano Res. 12, 1207–1215 (2019).
Afonin, K. A., Dobrovolskaia, M. A., Church, G. & Bathe, M. Opportunities, barriers, and a strategy for overcoming translational challenges to therapeutic nucleic acid nanotechnology. ACS Nano 14, 9221–9227 (2020).
Dobrovoskaia, M. A. & Bathe, M. Opportunities and challenges for the clinical translation of structured DNA assemblies as gene therapeutic delivery and vaccine vectors. Wiley Interdiscip. Nanomed. Nanobiotechnol. 13, e1657 (2021).
Afonin, K. A., Dobrovolskaia, M. A., Ke, W., Grodzinski, P. & Bathe, M. Critical review of nucleic acid nanotechnology to identify gaps and inform a strategy for accelerated clinical translation. Adv. Drug Deliv. Rev. 181, 114081 (2022).
Bujold, K. E., Lacroix, A. & Sleiman, H. F. DNA nanostructures at the interface with biology. Chem 4, 495–521 (2018).
Wamhoff, E. C. et al. Programming structured DNA assemblies to probe biophysical processes. Annu. Rev. Biophys. 48, 395–419 (2019).
Sacca, B. & Niemeyer, C. M. DNA origami: the art of folding DNA. Angew. Chem. Int. Ed. 51, 58–66 (2012).
Veneziano, R. et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat. Nanotechnol. 15, 716–723 (2020).
Hellmeier, J. et al. DNA origami demonstrate the unique stimulatory power of single pMHCs as T cell antigens. Proc. Natl Acad. Sci. USA 118, e2016857118 (2021).
Fang, T. et al. Spatial regulation of T-cell signaling by programmed death-ligand 1 on wireframe DNA origami flat sheets. ACS Nano 15, 3441–3452 (2021).
Kern, N., Dong, R., Douglas, S. M., Vale, R. D. & Morrissey, M. A. Tight nanoscale clustering of Fcgamma receptors using DNA origami promotes phagocytosis. eLife 10, e68311 (2021).
Dong, R. et al. DNA origami patterning of synthetic T cell receptors reveals spatial control of the sensitivity and kinetics of signal activation. Proc. Natl Acad. Sci. USA 118, e2109057118 (2021).
Rosier, B. et al. Proximity-induced caspase-9 activation on a DNA origami-based synthetic apoptosome. Nat. Catal. 3, 295–306 (2020).
Klein, W. P. et al. Enhanced catalysis from multienzyme cascades assembled on a DNA origami triangle. ACS Nano 13, 13677–13689 (2019).
Wang, D. et al. An addressable 2D heterogeneous nanoreactor to study the enzyme-catalyzed reaction at the interface. Small 13, 1700594 (2017).
Yang, Y. R. et al. 2D enzyme cascade network with efficient substrate channeling by swinging arms. ChemBioChem 19, 212–216 (2018).
Shaw, A. et al. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol. 14, 184–190 (2019).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Li, S. et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 (2018).
Wang, Z. et al. A tubular DNA nanodevice as a siRNA/chemo-drug co-delivery vehicle for combined cancer therapy. Angew. Chem. Int. Ed. 60, 2594–2598 (2021).
Liu, S. et al. A DNA nanodevice-based vaccine for cancer immunotherapy. Nat. Mater. 20, 421–430 (2021).
Dey, S. et al. DNA origami. Nat. Rev. Methods Primers 1, 13 (2021).
Angelin, A. et al. Multiscale origami structures as interface for cells. Angew. Chem. Int. Ed. 54, 15813–15817 (2015).
Bastings, M. M. C. et al. Modulation of the cellular uptake of DNA origami through control over mass and shape. Nano Lett. 18, 3557–3564 (2018).
Burns, J. R., Lamarre, B., Pyne, A. L. B., Noble, J. E. & Ryadnov, M. G. DNA origami inside-out viruses. ACS Synth. Biol. 7, 767–773 (2018).
Czogalla, A. et al. Amphipathic DNA origami nanoparticles to scaffold and deform lipid membrane vesicles. Angew. Chem. Int. Ed. 54, 6501–6505 (2015).
Czogalla, A., Kauert, D. J., Seidel, R., Schwille, P. & Petrov, E. P. DNA origami nanoneedles on freestanding lipid membranes as a tool to observe isotropic–nematic transition in two dimensions. Nano Lett. 15, 649–655 (2015).
Deng, Y. et al. Intracellular delivery of nanomaterials via an inertial microfluidic cell hydroporator. Nano Lett. 18, 2705–2710 (2018).
Fu, M. et al. Observation of intracellular interactions between DNA origami and lysosomes by the fluorescence localization method. Chem. Commun. 52, 9240–9242 (2016).
Funke, J. J., Ketterer, P., Lieleg, C., Korber, P. & Dietz, H. Exploring nucleosome unwrapping using DNA origami. Nano Lett. 16, 7891–7898 (2016).
Funke, J. J. et al. Uncovering the forces between nucleosomes using DNA origami. Sci. Adv. 2, e1600974 (2016).
Grome, M. W., Zhang, Z., Pincet, F. & Lin, C. Vesicle tubulation with self-assembling DNA nanosprings. Angew. Chem. Int. Ed. 57, 5330–5334 (2018).
Hirtz, M., Brglez, J., Fuchs, H. & Niemeyer, C. M. Selective binding of DNA origami on biomimetic lipid patches. Small 11, 5752–5758 (2015).
Hu, Y., Dominguez, C. M., Christ, S. & Niemeyer, C. M. Postsynthetic functionalization of DNA-nanocomposites with proteins yields bioinstructive matrices for cell culture applications. Angew. Chem. Int. Ed. 59, 19016–19020 (2020).
Ijas, H., Hakaste, I., Shen, B., Kostiainen, M. A. & Linko, V. Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo. ACS Nano 13, 5959–5967 (2019).
Jorge, A. F., Avino, A., Pais, A., Eritja, R. & Fabrega, C. DNA-based nanoscaffolds as vehicles for 5-fluoro-2’-deoxyuridine oligomers in colorectal cancer therapy. Nanoscale 10, 7238–7249 (2018).
Jusuk, I., Vietz, C., Raab, M., Dammeyer, T. & Tinnefeld, P. Super-resolution imaging conditions for enhanced yellow fluorescent protein (eYFP) demonstrated on DNA origami nanorulers. Sci. Rep. 5, 14075 (2015).
Khmelinskaia, A., Mucksch, J., Petrov, E. P., Franquelim, H. G. & Schwille, P. Control of membrane binding and diffusion of cholesteryl-modified DNA origami nanostructures by DNA spacers. Langmuir 34, 14921–14931 (2018).
Kocabey, S. et al. Cellular uptake of tile-assembled DNA nanotubes. Nanomaterials 5, 47–60 (2014).
Kosuri, P., Altheimer, B. D., Dai, M., Yin, P. & Zhuang, X. Rotation tracking of genome-processing enzymes using DNA origami rotors. Nature 572, 136–140 (2019).
Kramm, K. et al. DNA origami-based single-molecule force spectroscopy elucidates RNA polymerase III pre-initiation complex stability. Nat. Commun. 11, 2828 (2020).
List, J., Weber, M. & Simmel, F. C. Hydrophobic actuation of a DNA origami bilayer structure. Angew. Chem. Int. Ed. 53, 4236–4239 (2014).
Liu, J. et al. A tailored DNA nanoplatform for synergistic RNAi-/chemotherapy of multidrug-resistant tumors. Angew. Chem. Int. Ed. 57, 15486–15490 (2018).
Maezawa, T. et al. DNA density-dependent uptake of DNA origami-based two- or three-dimensional nanostructures by immune cells. Nanoscale 12, 14818–14824 (2020).
Mathur, D. & Henderson, E. R. Programmable DNA nanosystem for molecular interrogation. Sci. Rep. 6, 27413 (2016).
Ohtsuki, S. et al. Folding of single-stranded circular DNA into rigid rectangular DNA accelerates its cellular uptake. Nanoscale 11, 23416–23422 (2019).
Okholm, A. H. et al. Quantification of cellular uptake of DNA nanostructures by qPCR. Methods 67, 193–197 (2014).
Selnihhin, D., Sparvath, S. M., Preus, S., Birkedal, V. & Andersen, E. S. Multifluorophore DNA origami beacon as a biosensing platform. ACS Nano 12, 5699–5708 (2018).
Verma, V. et al. Using protein dimers to maximize the protein hybridization efficiency with multisite DNA origami scaffolds. PLoS ONE 10, e0137125 (2015).
Wu, S. et al. A quick-responsive DNA nanotechnology device for bio-molecular homeostasis regulation. Sci. Rep. 6, 31379 (2016).
Xing, C. et al. Spatial regulation of biomolecular interactions with a switchable trident-shaped DNA nanoactuator. ACS Appl. Mater. Interfaces 10, 32579–32587 (2018).
Zadegan, R. M. & Hughes, W. L. CAGE: chromatin analogous gene expression. ACS Synth. Biol. 6, 1800–1806 (2017).
Zhang, H. et al. DNA nanostructures coordinate gene silencing in mature plants. Proc. Natl Acad. Sci. USA 116, 7543–7548 (2019).
Schaffert, D. H. et al. Intracellular delivery of a planar DNA origami structure by the transferrin-receptor internalization pathway. Small 12, 2634–2640 (2016).
Kohman, R. E., Cha, S. S., Man, H. Y. & Han, X. Light-triggered release of bioactive molecules from DNA nanostructures. Nano Lett. 16, 2781–2785 (2016).
Journot, C. M. A., Ramakrishna, V., Wallace, M. I. & Turberfield, A. J. Modifying membrane morphology and interactions with DNA origami clathrin-mimic networks. ACS Nano 13, 9973–9979 (2019).
Silvester, E. et al. DNA origami signposts for identifying proteins on cell membranes by electron cryotomography. Cell 184, 1110–1121.e16 (2021).
Iwabuchi, S., Kawamata, I., Murata, S. & Nomura, S. M. A large, square-shaped, DNA origami nanopore with sealing function on a giant vesicle membrane. Chem. Commun. 57, 2990–2993 (2021).
Wang, Y. et al. DNA origami penetration in cell spheroid tissue models is enhanced by wireframe design. Adv. Mater. 33, e2008457 (2021).
Wu, X. et al. An RNA/DNA hybrid origami-based nanoplatform for efficient gene therapy. Nanoscale 13, 12848–12853 (2021).
Pal, S. & Rakshit, T. Folate-functionalized DNA origami for targeted delivery of doxorubicin to triple-negative breast cancer. Front. Chem. 9, 721105 (2021).
Smolkova, B. et al. Protein corona inhibits endosomal escape of functionalized DNA nanostructures in living cells. ACS Appl. Mater. Interfaces 13, 46375–46390 (2021).
Thomsen, R. P. et al. A large size-selective DNA nanopore with sensing applications. Nat. Commun. 10, 5655 (2019).
Huang, H. et al. Amphiphilic and biocompatible DNA origami-based emulsion formation and nanopore release for anti-melanogenesis therapy. Small 17, e2104831 (2021).
Hellmeier, J. et al. Strategies for the site-specific decoration of DNA origami nanostructures with functionally intact proteins. ACS Nano 15, 15057–15068 (2021).
Bell, N. A. & Keyser, U. F. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat. Nanotechnol. 11, 645–651 (2016).
Brglez, J., Ahmed, I. & Niemeyer, C. M. Photocleavable ligands for protein decoration of DNA nanostructures. Org. Biomol. Chem. 13, 5102–5104 (2015).
Iwaki, M., Wickham, S. F., Ikezaki, K., Yanagida, T. & Shih, W. M. A programmable DNA origami nanospring that reveals force-induced adjacent binding of myosin VI heads. Nat. Commun. 7, 13715 (2016).
Kielar, C. et al. Pharmacophore nanoarrays on DNA origami substrates as a single-molecule assay for fragment-based drug discovery. Angew. Chem. Int. Ed. 57, 14873–14877 (2018).
Kuzuya, A. et al. Allosteric control of nanomechanical DNA origami pinching devices for enhanced target binding. Chem. Commun. 53, 8276–8279 (2017).
Zhang, P. et al. Capturing transient antibody conformations with DNA origami epitopes. Nat. Commun. 11, 3114 (2020).
Zhang, P. et al. Quantitative measurement of spatial effects of DNA origami on molecular binding reactions detected using atomic force microscopy. ACS Appl. Mater. Interfaces 11, 21973–21981 (2019).
Liu, Y. et al. The effects of overhang placement and multivalency on cell labeling by DNA origami. Nanoscale 13, 6819–6828 (2021).
Wijesekara, P. et al. Accessing and assessing the cell-surface glycocalyx using DNA origami. Nano Lett. 21, 4765–4773 (2021).
Hoffecker, I. T. et al. Solution-controlled conformational switching of an anchored wireframe DNA nanostructure. Small 15, e1803628 (2019).
Eklund, A. S., Comberlato, A., Parish, I. A., Jungmann, R. & Bastings, M. M. C. Quantification of strand accessibility in biostable DNA origami with single-staple resolution. ACS Nano 15, 17668–17677 (2021).
Liu, K., Xu, C. & Liu, J. Regulation of cell binding and entry by DNA origami mediated spatial distribution of aptamers. J. Mater. Chem. B 8, 6802–6809 (2020).
Torelli, E. et al. Cotranscriptional folding of a bio-orthogonal fluorescent scaffolded RNA origami. ACS Synth. Biol. 9, 1682–1692 (2020).
Lu, Z., Wang, Y., Xu, D. & Pang, L. Aptamer-tagged DNA origami for spatially addressable detection of aflatoxin B1. Chem. Commun. 53, 941–944 (2017).
Zhao, S. et al. Efficient intracellular delivery of RNase A using DNA origami carriers. ACS Appl. Mater. Interfaces 11, 11112–11118 (2019).
Numajiri, K., Yamazaki, T., Kimura, M., Kuzuya, A. & Komiyama, M. Discrete and active enzyme nanoarrays on DNA origami scaffolds purified by affinity tag separation. J. Am. Chem. Soc. 132, 9937–9939 (2010).
Udomprasert, A. et al. Amyloid fibrils nucleated and organized by DNA origami constructions. Nat. Nanotechnol. 9, 537–541 (2014).
Sorensen, R. S. et al. Enzymatic ligation of large biomolecules to DNA. ACS Nano 7, 8098–8104 (2013).
Chen, X. et al. Aptamer-integrated scaffolds for biologically functional DNA origami structures. ACS Appl. Mater. Interfaces 13, 39711–39718 (2021).
Akinc, A. et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 26, 561–569 (2008).
Dahlman, J. E. et al. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl Acad. Sci. USA 114, 2060–2065 (2017).
Amir, Y. et al. Universal computing by DNA origami robots in a living animal. Nat. Nanotechnol. 9, 353–357 (2014).
Arnon, S. et al. Thought-controlled nanoscale robots in a living host. PLoS ONE 11, e0161227 (2016).
Chen, Y. et al. A synthetic light-driven substrate channeling system for precise regulation of enzyme cascade activity based on DNA origami. J. Am. Chem. Soc. 140, 8990–8996 (2018).
Derr, N. D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012).
Driller-Colangelo, A. R., Chau, K. W., Morgan, J. M. & Derr, N. D. Cargo rigidity affects the sensitivity of dynein ensembles to individual motor pausing. Cytoskeleton 73, 693–702 (2016).
Feng, Y., Hashiya, F., Hidaka, K., Sugiyama, H. & Endo, M. Direct observation of dynamic interactions between orientation-controlled nucleosomes in a DNA origami frame. Chemistry 26, 15282–15289 (2020).
Fisher, P. D. E. et al. A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement. ACS Nano 12, 1508–1518 (2018).
Fu, J. et al. Assembly of multienzyme complexes on DNA nanostructures. Nat. Protoc. 11, 2243–2273 (2016).
Fu, Y. et al. Single-step rapid assembly of DNA origami nanostructures for addressable nanoscale bioreactors. J. Am. Chem. Soc. 135, 696–702 (2013).
Fujita, K., Ohmachi, M., Ikezaki, K., Yanagida, T. & Iwaki, M. Direct visualization of human myosin II force generation using DNA origami-based thick filaments. Commun. Biol. 2, 437 (2019).
Ge, Z. et al. Constructing submonolayer DNA origami scaffold on gold electrode for wiring of redox enzymatic cascade pathways. ACS Appl. Mater. Interfaces 11, 13881–13887 (2019).
Grossi, G., Dalgaard Ebbesen Jepsen, M., Kjems, J. & Andersen, E. S. Control of enzyme reactions by a reconfigurable DNA nanovault. Nat. Commun. 8, 992 (2017).
Hahn, J., Chou, L. Y. T., Sorensen, R. S., Guerra, R. M. & Shih, W. M. Extrusion of RNA from a DNA-origami-based nanofactory. ACS Nano 14, 1550–1559 (2020).
Kaminska, I., Bohlen, J., Mackowski, S., Tinnefeld, P. & Acuna, G. P. Strong plasmonic enhancement of a single peridinin–chlorophyll a–protein complex on DNA origami-based optical antennas. ACS Nano 12, 1650–1655 (2018).
Ke, G. et al. Directional regulation of enzyme pathways through the control of substrate channeling on a DNA origami scaffold. Angew. Chem. Int. Ed. 55, 7483–7486 (2016).
Ke, Y., Meyer, T., Shih, W. M. & Bellot, G. Regulation at a distance of biomolecular interactions using a DNA origami nanoactuator. Nat. Commun. 7, 10935 (2016).
Ketterer, P. et al. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat. Commun. 9, 902 (2018).
Marth, G. et al. Precision templated bottom-up multiprotein nanoassembly through defined click chemistry linkage to DNA. ACS Nano 11, 5003–5010 (2017).
Matsuda, K. et al. Artificial smooth muscle model composed of hierarchically ordered microtubule asters mediated by DNA origami nanostructures. Nano Lett. 19, 3933–3938 (2019).
Rosier, B. et al. Incorporation of native antibodies and Fc-fusion proteins on DNA nanostructures via a modular conjugation strategy. Chem. Commun. 53, 7393–7396 (2017).
Shaw, A., Benson, E. & Hogberg, B. Purification of functionalized DNA origami nanostructures. ACS Nano 9, 4968–4975 (2015).
Shaw, A. et al. Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 11, 841–846 (2014).
Sommese, R. F. et al. Patterning protein complexes on DNA nanostructures using a GFP nanobody. Protein Sci. 25, 2089–2094 (2016).
Stanley, G. J. et al. Quantification of biomolecular dynamics inside real and synthetic nuclear pore complexes using time-resolved atomic force microscopy. ACS Nano 13, 7949–7956 (2019).
Stephanopoulos, N. et al. Immobilization and one-dimensional arrangement of virus capsids with nanoscale precision using DNA origami. Nano Lett. 10, 2714–2720 (2010).
Sun, L. et al. Real-time imaging of single-molecule enzyme cascade using a DNA origami raft. J. Am. Chem. Soc. 139, 17525–17532 (2017).
Vogele, K., List, J., Simmel, F. C. & Pirzer, T. Enhanced efficiency of an enzyme cascade on DNA-activated silica surfaces. Langmuir 34, 14780–14786 (2018).
Wang, D. et al. A switchable DNA origami nanochannel for regulating molecular transport at the nanometer scale. Nanoscale 8, 3944–3948 (2016).
Weinhold, E. & Chakraborty, B. DNA modification and visualization on an origami-based enzyme nano-factory. Nanoscale 13, 2465–2471 (2021).
Williams, N. D. et al. DNA-origami-based fluorescence brightness standards for convenient and fast protein counting in live cells. Nano Lett. 20, 8890–8896 (2020).
Xu, W. et al. A programmable DNA origami platform to organize SNAREs for membrane fusion. J. Am. Chem. Soc. 138, 4439–4447 (2016).
Xu, Y. et al. Single-molecule studies of allosteric inhibition of individual enzyme on a DNA origami reactor. J. Phys. Chem. Lett. 9, 6786–6794 (2018).
Zanacchi, F. C. et al. A DNA origami platform for quantifying protein copy number in super-resolution. Nat. Methods 14, 789–792 (2017).
Zhao, Z. et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 7, 10619 (2016).
Zhou, J. et al. A new biochromatography model based on DNA origami assembled PPARgamma: construction and evaluation. Anal. Bioanal. Chem. 409, 3059–3065 (2017).
Rai, A., Vang, D., Ritt, M. & Sivaramakrishnan, S. Dynamic multimerization of Dab2-Myosin VI complexes regulates cargo processivity while minimizing cortical actin reorganization. J. Biol. Chem. 296, 100232 (2021).
Abe, K., Sugiyama, H. & Endo, M. Construction of an optically controllable CRISPR-Cas9 system using a DNA origami nanostructure. Chem. Commun. 57, 5594–5596 (2021).
Berger, R. M. L. et al. Nanoscale FasL organization on DNA origami to decipher apoptosis signal activation in cells. Small 17, e2101678 (2021).
Cremers, G. A. O. et al. Determinants of ligand-functionalized DNA nanostructure–cell interactions. J. Am. Chem. Soc. 143, 10131–10142 (2021).
Shen, Q. et al. DNA-origami nanotrap for studying the selective barriers formed by phenylalanine-glycine-rich nucleoporins. J. Am. Chem. Soc. 143, 12294–12303 (2021).
Sigl, C. et al. Programmable icosahedral shell system for virus trapping. Nat. Mater. 20, 1281–1289 (2021).
Sprengel, A. et al. Tailored protein encapsulation into a DNA host using geometrically organized supramolecular interactions. Nat. Commun. 8, 14472 (2017).
Akbari, E. et al. Engineering cell surface function with DNA origami. Adv. Mater. 29, 1703632 (2017).
Chopra, A., Krishnan, S. & Simmel, F. C. Electrotransfection of polyamine folded DNA origami structures. Nano Lett. 16, 6683–6690 (2016).
Dai, M., Jungmann, R. & Yin, P. Optical imaging of individual biomolecules in densely packed clusters. Nat. Nanotechnol. 11, 798–807 (2016).
Dutta, P. K. et al. Programmable multivalent DNA-origami tension probes for reporting cellular traction forces. Nano Lett. 18, 4803–4811 (2018).
Johnson-Buck, A., Jiang, S., Yan, H. & Walter, N. G. DNA-cholesterol barges as programmable membrane-exploring agents. ACS Nano 8, 5641–5649 (2014).
Kaplan, C. et al. Absolute arrangement of subunits in cytoskeletal septin filaments in cells measured by fluorescence microscopy. Nano Lett. 15, 3859–3864 (2015).
Mucksch, J. et al. Quantifying reversible surface binding via surface-integrated fluorescence correlation spectroscopy. Nano Lett. 18, 3185–3192 (2018).
Rahman, M. A. et al. Systemic delivery of Bc12-targeting siRNA by DNA nanoparticles suppresses cancer cell growth. Angew. Chem. Int. Ed. 56, 16023–16027 (2017).
Tohgasaki, T. et al. A photocaged DNA nanocapsule for controlled unlocking and opening inside the cell. Bioconjug. Chem. 30, 1860–1863 (2019).
Wang, P. et al. Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells. J. Am. Chem. Soc. 140, 2478–2484 (2018).
Wiraja, C. et al. Framework nucleic acids as programmable carrier for transdermal drug delivery. Nat. Commun. 10, 1147 (2019).
Ge, Z. et al. DNA origami-enabled engineering of ligan–drug conjugates for targeted drug delivery. Small 16, e1904857 (2020).
Franquelim, H. G., Dietz, H. & Schwille, P. Reversible membrane deformations by straight DNA origami filaments. Soft Matter 17, 276–287 (2021).
Lin, P., Dinh, H., Nakata, E. & Morii, T. Dynamic shape transformation of a DNA scaffold applied for an enzyme nanocarrier. Front. Chem. 9, 697857 (2021).
Jahnke, K. et al. Proton gradients from light-harvesting E. coli control DNA assemblies for synthetic cells. Nat. Commun. 12, 3967 (2021).
Fragasso, A. et al. Reconstitution of ultrawide DNA origami pores in liposomes for transmembrane transport of macromolecules. ACS Nano 15, 12768–12779 (2021).
Liu, J. et al. Reconstructing Soma-Soma synapse-like vesicular exocytosis with DNA origami. ACS Cent. Sci. 7, 1400–1407 (2021).
Daems, D. et al. Controlling the bioreceptor spatial distribution at the nanoscale for single molecule counting in microwell arrays. ACS Sens. 4, 2327–2335 (2019).
Pan, Q. et al. Aptamer-functionalized DNA origami for targeted codelivery of antisense oligonucleotides and doxorubicin to enhance therapy in drug-resistant cancer cells. ACS Appl. Mater. Interfaces 12, 400–409 (2020).
Raveendran, M., Lee, A. J., Sharma, R., Walti, C. & Actis, P. Rational design of DNA nanostructures for single molecule biosensing. Nat. Commun. 11, 4384 (2020).
Rutten, I., Daems, D. & Lammertyn, J. Boosting biomolecular interactions through DNA origami nano-tailored biosensing interfaces. J. Mater. Chem. B 8, 3606–3615 (2020).
Sun, P. C. et al. Site-specific anchoring aptamer C2NP on DNA origami nanostructures for cancer treatment. RSC Adv. 8, 26300–26308 (2018).
Yang, L. et al. An intelligent DNA nanorobot for autonomous anticoagulation. Angew. Chem. Int. Ed. 59, 17697–17704 (2020).
Zhao, S. et al. A DNA origami-based aptamer nanoarray for potent and reversible anticoagulation in hemodialysis. Nat. Commun. 12, 358 (2021).
Xu, F. et al. Profiling and regulating proteins that adsorb to DNA materials in human serum. Anal. Chem. 93, 8671–8679 (2021).
Liu, F. et al. A tetrahedral DNA nanorobot with conformational change in response to molecular trigger. Nanoscale 13, 15552–15559 (2021).
Zhang, T. et al. Construction of a reconfigurable DNA nanocage for encapsulating a TMV disk. Chem. Commun. 55, 8951–8954 (2019).
Zhou, K., Ke, Y. & Wang, Q. Selective in situ assembly of viral protein onto DNA origami. J. Am. Chem. Soc. 140, 8074–8077 (2018).
Zhou, K., Zhou, Y., Pan, V., Wang, Q. & Ke, Y. Programming dynamic assembly of viral proteins with DNA origami. J. Am. Chem. Soc. 142, 5929–5932 (2020).
Xu, T. et al. DNA origami frameworks enabled self-protective siRNA delivery for dual enhancement of chemo-photothermal combination therapy. Small 17, e2101780 (2021).
Buchberger, A., Simmons, C. R., Fahmi, N. E., Freeman, R. & Stephanopoulos, N. Hierarchical assembly of nucleic acid/coiled-coil peptide nanostructures. J. Am. Chem. Soc. 142, 1406–1416 (2020).
Hawkes, W. et al. Probing the nanoscale organisation and multivalency of cell surface receptors: DNA origami nanoarrays for cellular studies with single-molecule control. Faraday Discuss. 219, 203–219 (2019).
Huang, D., Patel, K., Perez-Garrido, S., Marshall, J. F. & Palma, M. DNA origami nanoarrays for multivalent investigations of cancer cell spreading with nanoscale spatial resolution and single-molecule control. ACS Nano 13, 728–736 (2019).
Karna, D. et al. Chemo-mechanical modulation of cell motions using DNA nanosprings. Bioconjug. Chem. 32, 311–317 (2021).
Wang, Y., Baars, I., Fordos, F. & Hogberg, B. Clustering of death receptor for apoptosis using nanoscale patterns of peptides. ACS Nano 15, 9614–9626 (2021).
Bian, X., Zhang, Z., Xiong, Q., De Camilli, P. & Lin, C. A programmable DNA-origami platform for studying lipid transfer between bilayers. Nat. Chem. Biol. 15, 830–837 (2019).
Dong, Y. et al. Folding DNA into a lipid-conjugated nanobarrel for controlled reconstitution of membrane proteins. Angew. Chem. Int. Ed. 57, 2072–2076 (2018).
Yang, Y. et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat. Chem. 8, 476–483 (2016).
Zhang, Z. & Chapman, E. R. Programmable nanodisc patterning by DNA origami. Nano Lett. 20, 6032–6037 (2020).
Zhang, Z., Yang, Y., Pincet, F., Llaguno, M. C. & Lin, C. Placing and shaping liposomes with reconfigurable DNA nanocages. Nat. Chem. 9, 653–659 (2017).
Zhao, Z. et al. DNA-corralled nanodiscs for the structural and functional characterization of membrane proteins and viral entry. J. Am. Chem. Soc. 140, 10639–10643 (2018).
Gopfrich, K. et al. Large-conductance transmembrane porin made from DNA origami. ACS Nano 10, 8207–8214 (2016).
Du, Y. et al. DNA-nanostructure-gold-nanorod hybrids for enhanced in vivo optoacoustic imaging and photothermal therapy. Adv. Mater. 28, 10000–10007 (2016).
Jiang, D. et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat. Biomed. Eng. 2, 865–877 (2018).
Jiang, Q. et al. A self-assembled DNA origami–gold nanorod complex for cancer theranostics. Small 11, 5134–5141 (2015).
Kaur, V., Tanwar, S., Kaur, G. & Sen, T. DNA-origami-based assembly of Au@Ag nanostar dimer nanoantennas for label-free sensing of pyocyanin. ChemPhysChem 22, 160–167 (2021).
Meyer, T. A., Zhang, C., Bao, G. & Ke, Y. Programmable assembly of iron oxide nanoparticles using DNA origami. Nano Lett. 20, 2799–2805 (2020).
Song, L. et al. DNA origami/gold nanorod hybrid nanostructures for the circumvention of drug resistance. Nanoscale 9, 7750–7754 (2017).
Takenaka, T. et al. Photoresponsive DNA nanocapsule having an open/close system for capture and release of nanomaterials. Chemistry 20, 14951–14954 (2014).
Agarwal, N. P., Matthies, M., Gur, F. N., Osada, K. & Schmidt, T. L. Block copolymer micellization as a protection strategy for DNA origami. Angew. Chem. Int. Ed. 56, 5460–5464 (2017).
Liu, J. et al. A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy. Nano Lett. 18, 3328–3334 (2018).
Kim, Y. & Yin, P. Enhancing biocompatible stability of DNA nanostructures using dendritic oligonucleotides and brick motifs. Angew. Chem. Int. Ed. 59, 700–703 (2020).
Nakata, E. et al. Zinc-finger proteins for site-specific protein positioning on DNA-origami structures. Angew. Chem. Int. Ed. 51, 2421–2424 (2012).
Sagredo, S. et al. Orthogonal protein assembly on DNA nanostructures using relaxases. Angew. Chem. Int. Ed. 55, 4348–4352 (2016).
Kurokawa, T. et al. DNA origami scaffolds as templates for functional tetrameric Kir3 K(+) channels. Angew. Chem. Int. Ed. 57, 2586–2591 (2018).
Takusagawa, M. et al. HBD1 protein with a tandem repeat of two HMG-box domains is a DNA clip to organize chloroplast nucleoids in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 118, e2021053118 (2021).
Aksel, T., Yu, Z., Cheng, Y. & Douglas, S. M. Molecular goniometers for single-particle cryo-electron microscopy of DNA-binding proteins. Nat. Biotechnol. 39, 378–386 (2021).
Kuzuya, A. et al. Precisely programmed and robust 2D streptavidin nanoarrays by using periodical nanometer-scale wells embedded in DNA origami assembly. ChemBioChem 10, 1811–1815 (2009).
Kuzyk, A., Laitinen, K. T. & Torma, P. DNA origami as a nanoscale template for protein assembly. Nanotechnology 20, 235305 (2009).
Le, J. V. et al. Probing nucleosome stability with a DNA origami nanocaliper. ACS Nano 10, 7073–7084 (2016).
Linko, V., Eerikainen, M. & Kostiainen, M. A. A modular DNA origami-based enzyme cascade nanoreactor. Chem. Commun. 51, 5351–5354 (2015).
Mallik, L. et al. Electron microscopic visualization of protein assemblies on flattened DNA origami. ACS Nano 9, 7133–7141 (2015).
Numajiri, K., Kimura, M., Kuzuya, A. & Komiyama, M. Stepwise and reversible nanopatterning of proteins on a DNA origami scaffold. Chem. Commun. 46, 5127–5129 (2010).
Ora, A. et al. Cellular delivery of enzyme-loaded DNA origami. Chem. Commun. 52, 14161–14164 (2016).
Timm, C. & Niemeyer, C. M. Assembly and purification of enzyme-functionalized DNA origami structures. Angew. Chem. Int. Ed. 4, 6745–6750 (2015).
Yamazaki, T., Heddle, J. G., Kuzuya, A. & Komiyama, M. Orthogonal enzyme arrays on a DNA origami scaffold bearing size-tunable wells. Nanoscale 6, 9122–9126 (2014).
Yan, J. et al. Novel rolling circle amplification and DNA origami-based DNA belt-involved signal amplification assay for highly sensitive detection of prostate-specific antigen (PSA). ACS Appl. Mater. Interfaces 6, 20372–20377 (2014).
Pedersen, R. O., Loboa, E. G. & LaBean, T. H. Sensitization of transforming growth factor-beta signaling by multiple peptides patterned on DNA nanostructures. Biomacromolecules 14, 4157–4160 (2013).
Zhang, Q. et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 8, 6633–6643 (2014).
Breitz, H. B. et al. Clinical optimization of pretargeted radioimmunotherapy with antibody–streptavidin conjugate and 90Y-DOTA-biotin. J. Nucl. Med. 41, 131–140 (2000).
Meyer, D. L. et al. Reduced antibody response to streptavidin through site-directed mutagenesis. Protein Sci. 10, 491–503 (2001).
Mao, X. et al. Directing curli polymerization with DNA origami nucleators. Nat. Commun. 10, 1395 (2019).
Shen, W., Zhong, H., Neff, D. & Norton, M. L. NTA directed protein nanopatterning on DNA origami nanoconstructs. J. Am. Chem. Soc. 131, 6660–6661 (2009).
Brglez, J., Nikolov, P., Angelin, A. & Niemeyer, C. M. Designed intercalators for modification of DNA origami surface properties. Chemistry 21, 9440–9446 (2015).
Halley, P. D. et al. Daunorubicin-loaded DNA origami nanostructures circumvent drug-resistance mechanisms in a leukemia model. Small 12, 308–320 (2016).
Jiang, Q. et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 134, 13396–13403 (2012).
Miller, H. L. et al. Biophysical characterisation of DNA origami nanostructures reveals inaccessibility to intercalation binding sites. Nanotechnology 31, 235605 (2020).
Palazzolo, S. et al. An effective multi-stage liposomal DNA origami nanosystem for in vivo cancer therapy. Cancers 11, 1997 (2019).
Palazzolo, S. et al. Proof-of-concept multistage biomimetic liposomal DNA origami nanosystem for the remote loading of doxorubicin. ACS Med. Chem. Lett. 10, 517–521 (2019).
Zeng, Y. et al. Time-lapse live cell imaging to monitor doxorubicin release from DNA origami nanostructures. J. Mater. Chem. B 6, 1605–1612 (2018).
Zhao, Y. X. et al. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 6, 8684–8691 (2012).
Zhuang, X. et al. A photosensitizer-loaded DNA origami nanosystem for photodynamic therapy. ACS Nano 10, 3486–3495 (2016).
Ijas, H. et al. Unraveling the interaction between doxorubicin and DNA origami nanostructures for customizable chemotherapeutic drug release. Nucleic Acids Res. 49, 3048–3062 (2021).
Mikkila, J. et al. Virus-encapsulated DNA origami nanostructures for cellular delivery. Nano Lett. 14, 2196–2200 (2014).
Auvinen, H. et al. Protein coating of DNA nanostructures for enhanced stability and immunocompatibility. Adv. Healthc. Mater. 6, 1700692 (2017).
Hernandez-Garcia, A. et al. Precise coating of a wide range of DNA templates by a protein polymer with a DNA binding domain. ACS Nano 11, 144–152 (2017).
Kopatz, I. et al. Packaging of DNA origami in viral capsids. Nanoscale 11, 10160–10166 (2019).
Xu, X. et al. Cationic albumin encapsulated DNA origami for enhanced cellular transfection and stability. Materials 12, 949 (2019).
Wang, Y. et al. A nanoscale DNA force spectrometer capable of applying tension and compression on biomolecules. Nucleic Acids Res. 49, 8987–8999 (2021).
Shen, X. et al. Visualization of the intracellular location and stability of DNA origami with a label-free fluorescent probe. Chem. Commun. 48, 11301–11303 (2012).
Kiviaho, J. K. et al. Cationic polymers for DNA origami coating — examining their binding efficiency and tuning the enzymatic reaction rates. Nanoscale 8, 11674–11680 (2016).
Ponnuswamy, N. et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 8, 15654 (2017).
Ahmadi, Y., De Llano, E. & Barisic, I. (Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures. Nanoscale 10, 7494–7504 (2018).
Ahmadi, Y. & Barisic, I. Gene-therapy inspired polycation coating for protection of DNA origami nanostructures. J. Vis. Exp. https://doi.org/10.3791/58771 (2019).
Wang, S. T. et al. DNA origami protection and molecular interfacing through engineered sequence-defined peptoids. Proc. Natl Acad. Sci. USA 117, 6339–6348 (2020).
Anastassacos, F. M., Zhao, Z., Zeng, Y. & Shih, W. M. Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation. J. Am. Chem. Soc. 142, 3311–3315 (2020).
Estrich, N. A., Hernandez-Garcia, A., de Vries, R. & LaBean, T. H. Engineered diblock polypeptides improve DNA and gold solubility during molecular assembly. ACS Nano 11, 831–842 (2017).
Julin, S., Nonappa, Shen, B., Linko, V. & Kostiainen, M. A. DNA-origami-templated growth of multilamellar lipid assemblies. Angew. Chem. Int. Ed. 60, 827–833 (2021).
Shaukat, A. et al. Phthalocyanine–DNA origami complexes with enhanced stability and optical properties. Chem. Commun. 56, 7341–7344 (2020).
Sacca, B. et al. Orthogonal protein decoration of DNA origami. Angew. Chem. Int. Ed. 49, 9378–9383 (2010).
Masubuchi, T. et al. Construction of integrated gene logic-chip. Nat. Nanotechnol. 13, 933–940 (2018).
Nguyen, T. M., Nakata, E., Saimura, M., Dinh, H. & Morii, T. Design of modular protein tags for orthogonal covalent bond formation at specific DNA sequences. J. Am. Chem. Soc. 139, 8487–8496 (2017).
Angelin, A., Kassel, O., Rastegar, S., Strahle, U. & Niemeyer, C. M. Protein-functionalized DNA nanostructures as tools to control transcription in zebrafish embryos. ChemistryOpen 6, 33–39 (2017).
Nakata, E., Dinh, H., Ngo, T. A., Saimura, M. & Morii, T. A modular zinc finger adaptor accelerates the covalent linkage of proteins at specific locations on DNA nanoscaffolds. Chem. Commun. 51, 1016–1019 (2015).
Burgahn, T., Garrecht, R., Rabe, K. S. & Niemeyer, C. M. Solid-phase synthesis and purification of protein–DNA origami nanostructures. Chemistry 25, 3483–3488 (2019).
Kossmann, K. J. et al. A rationally designed connector for assembly of protein-functionalized DNA nanostructures. ChemBioChem 17, 1102–1106 (2016).
Zhang, Z. et al. Tuning the reactivity of a substrate for SNAP-tag expands its application for recognition-driven DNA–protein conjugation. Chemistry 27, 18118–18128 (2021).
Kroll, S., Rabe, K. S. & Niemeyer, C. M. An orthogonal covalent connector system for the efficient assembly of enzyme cascades on DNA nanostructures. Small 17, e2105095 (2021).
Stephanopoulos, N. & Francis, M. B. Choosing an effective protein bioconjugation strategy. Nat. Chem. Biol. 7, 876–884 (2011).
Ouyang, X. et al. Docking of antibodies into the cavities of DNA origami structures. Angew. Chem. Int. Ed. 56, 14423–14427 (2017).
Voigt, N. V. et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 5, 200–203 (2010).
Kolb, H. C., Finn, M. G. & Sharpless, K. B. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 40, 2004–2021 (2001).
Raniolo, S. et al. Cellular uptake of covalent and non-covalent DNA nanostructures with different sizes and geometries. Nanoscale 11, 10808–10818 (2019).
Dommerholt, J., Rutjes, F. & van Delft, F. L. Strain-promoted 1,3-dipolar cycloaddition of cycloalkynes and organic azides. Top. Curr. Chem. 374, 16 (2016).
McKay, C. S. & Finn, M. G. Click chemistry in complex mixtures: bioorthogonal bioconjugation. Chem. Biol. 21, 1075–1101 (2014).
Lin, Z., Xiong, Y., Xiang, S. & Gang, O. Controllable covalent-bound nanoarchitectures from DNA frames. J. Am. Chem. Soc. 141, 6797–6801 (2019).
Heck, C. et al. Label as you fold: methyltransferase-assisted functionalization of DNA nanostructures. Nanoscale 12, 20287–20291 (2020).
Knappe, G. A., Wamhoff, E. C., Read, B. J., Irvine, D. J. & Bathe, M. In situ covalent functionalization of DNA origami virus-like particles. ACS Nano 15, 14316–14322 (2021).
Oliveira, B. L., Guo, Z. & Bernardes, G. J. L. Inverse electron demand Diels–Alder reactions in chemical biology. Chem. Soc. Rev. 46, 4895–4950 (2017).
Zhang, J. X. et al. Predicting DNA hybridization kinetics from sequence. Nat. Chem. 10, 91–98 (2018).
Srisa-Art, M., Dyson, E. C., deMello, A. J. & Edel, J. B. Monitoring of real-time streptavidin–biotin binding kinetics using droplet microfluidics. Anal. Chem. 80, 7063–7067 (2008).
Kanekiyo, M. et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 20, 362–372 (2019).
Cohen, A. A. et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741 (2021).
Wagenbauer, K. F. et al. How we make DNA origami. ChemBioChem 18, 1873–1885 (2017).
Cervantes-Salguero, K., Freeley, M., Chavez, J. L. & Palma, M. Single-molecule DNA origami aptasensors for real-time biomarker detection. J. Mater. Chem. B 8, 6352–6356 (2020).
Lin, C., Perrault, S. D., Kwak, M., Graf, F. & Shih, W. M. Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 41, e40 (2013).
Stahl, E., Martin, T. G., Praetorius, F. & Dietz, H. Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. 53, 12735–12740 (2014).
Fahrner, R. L. et al. Industrial purification of pharmaceutical antibodies: development, operation, and validation of chromatography processes. Biotechnol. Genet. Eng. Rev. 18, 301–327 (2001).
Johnston, A. & Adcock, W. The use of chromatography to manufacture purer and safer plasma products. Biotechnol. Genet. Eng. Rev. 17, 37–70 (2000).
Mathur, D. & Medintz, I. L. Analyzing DNA nanotechnology: a call to arms for the analytical chemistry community. Anal. Chem. 89, 2646–2663 (2017).
Mehnert, W. & Mader, K. Solid lipid nanoparticles: production, characterization and applications. Adv. Drug Deliv. Rev. 47, 165–196 (2001).
Kretschy, N., Sack, M. & Somoza, M. M. Sequence-dependent fluorescence of Cy3- and Cy5-labeled double-stranded DNA. Bioconjug. Chem. 27, 840–848 (2016).
Funke, J. J. & Dietz, H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 11, 47–52 (2016).
Strauss, M. T., Schueder, F., Haas, D., Nickels, P. C. & Jungmann, R. Quantifying absolute addressability in DNA origami with molecular resolution. Nat. Commun. 9, 1600 (2018).
Bertosin, E. et al. Cryo-electron microscopy and mass analysis of oligolysine-coated DNA nanostructures. ACS Nano 15, 9391–9403 (2021).
van Dyck, J. F. et al. Sizing up DNA nanostructure assembly with native mass spectrometry and ion mobility. Nat. Commun. 13, 3610 (2022).
Jain, S. S. & Tullius, T. D. Footprinting protein–DNA complexes using the hydroxyl radical. Nat. Protoc. 3, 1092–1100 (2008).
Weeks, K. M. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 20, 295–304 (2010).
Zubradt, M. et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82 (2017).
Parsons, M. F. et al. 3D RNA-scaffolded wireframe origami. Nat. Commun. (in the press).
Acknowledgements
G.A.K., E.-C.W. and M.B. were supported by NIH R01-Al162307, NIH R21-EB026008-S1, ONR N00014-21-1-4013, ONR N00014-20-1-2084, NSF CCF-1956054, ARO ISN W911NF-13-D-0001 and ARO ICB Subaward KK1954. G.A.K. was additionally supported by the National Science Foundation under a Graduate Research Fellowship 2389237.
Author information
Authors and Affiliations
Contributions
G.A.K. and M.B. outlined the article. G.A.K. wrote the article. G.A.K., E.-C.W. and M.B. edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Materials thanks Veikko Linko, Sandra Kröll, Jibin Abraham Punnoose and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Knappe, G.A., Wamhoff, EC. & Bathe, M. Functionalizing DNA origami to investigate and interact with biological systems. Nat Rev Mater 8, 123–138 (2023). https://doi.org/10.1038/s41578-022-00517-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41578-022-00517-x
This article is cited by
-
Twisted moiré conductive thermal metasurface
Nature Communications (2024)
-
Enhancing antibody responses by multivalent antigen display on thymus-independent DNA origami scaffolds
Nature Communications (2024)
-
Imaging DNA origami by fluorescence in situ hybridization
Nature Nanotechnology (2024)